library(tidyverse)
library(scales)
library(janitor)
library(colorspace)
aqi_levels <- tribble(
~aqi_min, ~aqi_max, ~color, ~level,
0, 50, "#D8EEDA", "Good",
51, 100, "#F1E7D4", "Moderate",
101, 150, "#F8E4D8", "Unhealthy for sensitive groups",
151, 200, "#FEE2E1", "Unhealthy",
201, 300, "#F4E3F7", "Very unhealthy",
301, 400, "#F9D0D4", "Hazardous"
)AE: Air quality index in Syracuse, NY
Suggested answers
Packages
Import and clean Syracuse 2023 data
syr_2023 <- read_csv(file = "data/aqi-syracuse/ad_aqi_tracker_data-2023.csv")Rows: 365 Columns: 11
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (7): Date, Main Pollutant, Site Name, Site ID, Source, Date of 20-year H...
dbl (4): AQI Value, 20-year High (2000-2019), 20-year Low (2000-2019), 5-yea...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.syr_2023 <- syr_2023 |>
clean_names() |>
mutate(date = mdy(date))
syr_2023# A tibble: 365 × 11
date aqi_value main_pollutant site_name site_id source
<date> <dbl> <chr> <chr> <chr> <chr>
1 2023-01-01 38 PM2.5 EAST SYRACUSE 36-067-1015 AQS
2 2023-01-02 48 PM2.5 EAST SYRACUSE 36-067-1015 AQS
3 2023-01-03 49 PM2.5 EAST SYRACUSE 36-067-1015 AQS
4 2023-01-04 22 PM2.5 EAST SYRACUSE 36-067-1015 AQS
5 2023-01-05 33 PM2.5 EAST SYRACUSE 36-067-1015 AQS
6 2023-01-06 33 PM2.5 EAST SYRACUSE 36-067-1015 AQS
7 2023-01-07 30 PM2.5 EAST SYRACUSE 36-067-1015 AQS
8 2023-01-08 28 PM2.5 EAST SYRACUSE 36-067-1015 AQS
9 2023-01-09 50 PM2.5 EAST SYRACUSE 36-067-1015 AQS
10 2023-01-10 28 Ozone FULTON 36-075-0003 AQS
# ℹ 355 more rows
# ℹ 5 more variables: x20_year_high_2000_2019 <dbl>,
# x20_year_low_2000_2019 <dbl>, x5_year_average_2015_2019 <dbl>,
# date_of_20_year_high <chr>, date_of_20_year_low <chr>Recreate the plot
Basic line chart
Demo: Draw a simple line chart of the AQI in Syracuse for 2023.
Add color shading
Your turn: Add color shading to the plot based on the AQI guide. The color palette does not need to match the specific colors in the table yet.
You can use geom_rect() to draw rectangles on a plot. Remember the layered grammar of graphics and arrange the geom_*() in an appropriate order.
# draw the graph
syr_2023 |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
# shade in background with colors based on AQI guide
geom_rect(
data = aqi_levels,
aes(
ymin = aqi_min, ymax = aqi_max,
xmin = as.Date(-Inf), xmax = as.Date(Inf),
x = NULL, y = NULL, fill = color
)
) +
# plot the AQI in Syracuse
geom_line(linewidth = 1, alpha = 0.5)Use scale_fill_identity()
Your turn: Use the hexidecimal colors from the dataset for the color palette.
# draw the graph
syr_2023 |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
# shade in background with colors based on AQI guide
geom_rect(
data = aqi_levels,
aes(
ymin = aqi_min, ymax = aqi_max,
xmin = as.Date(-Inf), xmax = as.Date(Inf),
x = NULL, y = NULL, fill = color
)
) +
# use the hexidecimal colors from the dataset for the palette
scale_fill_identity() +
# plot the AQI in Syracuse
geom_line(linewidth = 1, alpha = 0.5)Label each AQI category on the chart
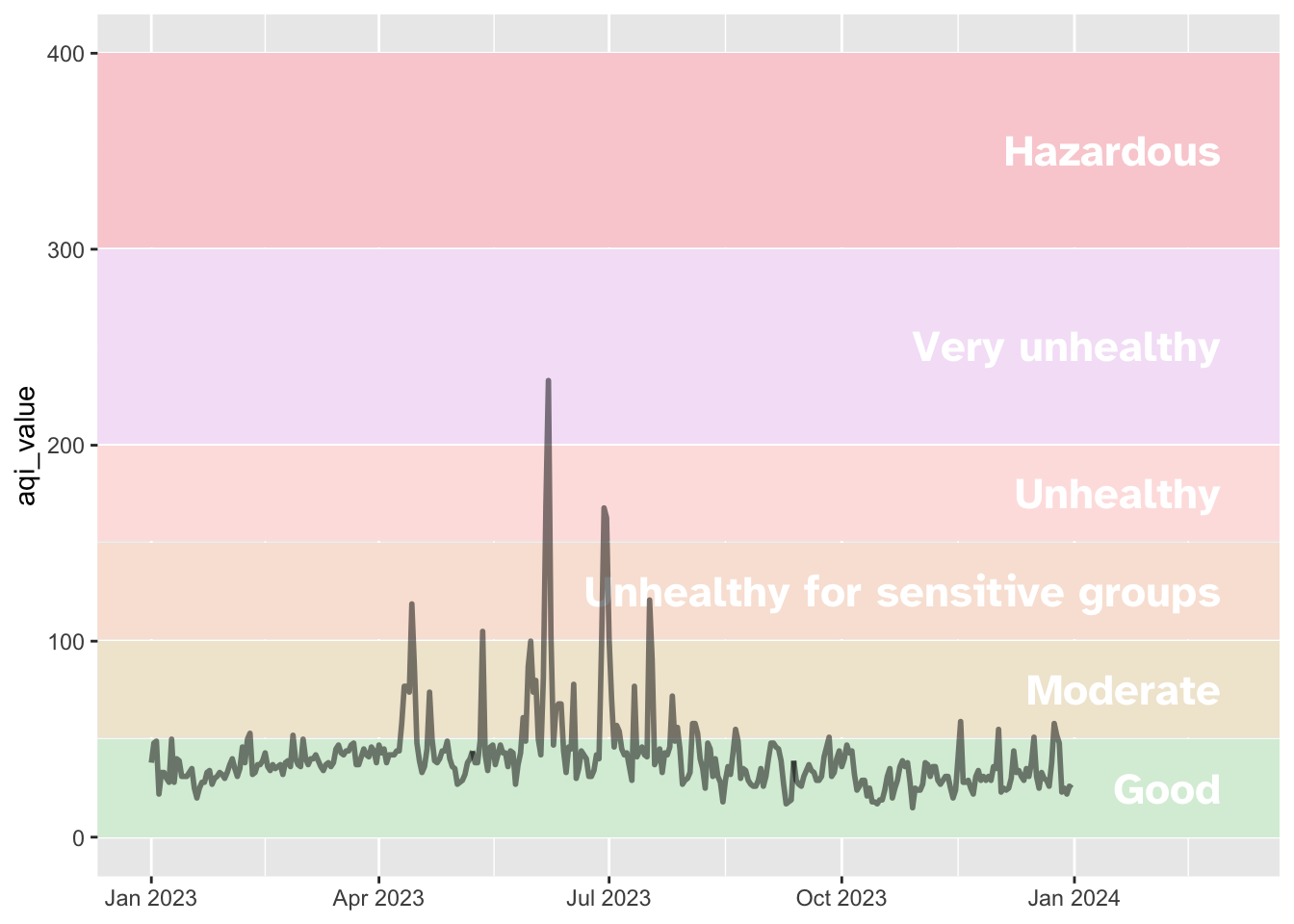
Your turn: Incorporate text labels for each AQI value directly into the graph. To accomplish this, you need to”
- Calculate the midpoint AQI value for each category
- Add a
geom_text()layer to the plot with the AQI values positioned at the midpoint on the \(y\)-axis
Extend the range of the \(x\)-axis to provide more horizontal space for the AQI category labels without interfering with the trend line.
aqi_levels <- aqi_levels |>
mutate(aqi_mid = ((aqi_min + aqi_max) / 2))
# draw the graph
syr_2023 |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
# shade in background with colors based on AQI guide
geom_rect(
data = aqi_levels,
aes(
ymin = aqi_min, ymax = aqi_max,
xmin = as.Date(-Inf), xmax = as.Date(Inf),
x = NULL, y = NULL, fill = color
)
) +
# use the hexidecimal colors from the dataset for the palette
scale_fill_identity() +
# format the x-axis for dates
scale_x_date(
name = NULL, date_labels = "%b %Y",
limits = c(ymd("2023-01-01"), ymd("2024-03-01"))
) +
# add text labels for each AQI category
geom_text(
data = aqi_levels,
aes(x = ymd("2024-02-28"), y = aqi_mid, label = level),
hjust = 1, size = 6, fontface = "bold", color = "white",
family = "Atkinson Hyperlegible"
) +
# plot the AQI in Syracuse
geom_line(linewidth = 1, alpha = 0.5)Add labels
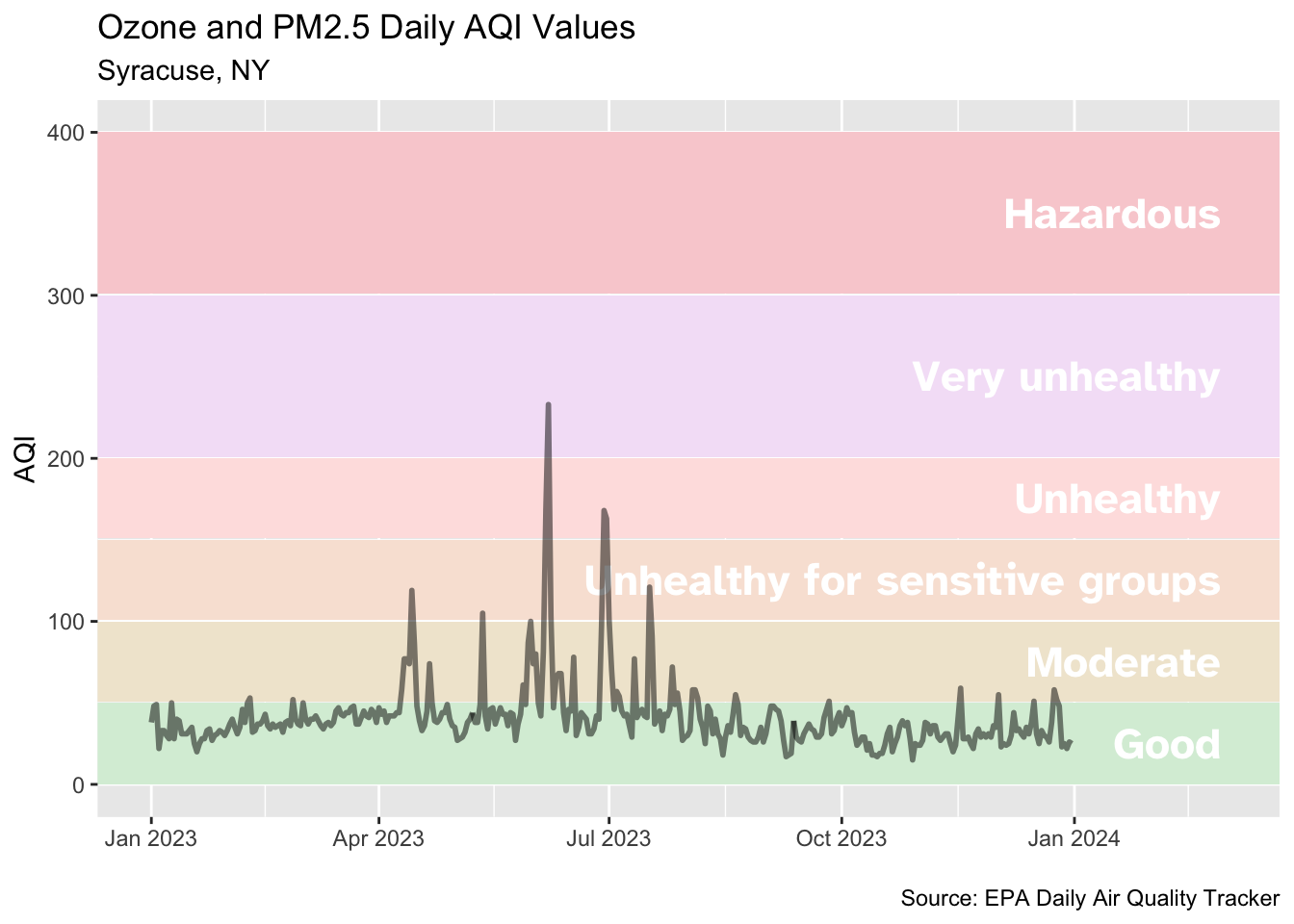
Demo: Add a meaningful title, axis labels, caption, etc.
# draw the graph
syr_2023 |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
# shade in background with colors based on AQI guide
geom_rect(
data = aqi_levels,
aes(
ymin = aqi_min, ymax = aqi_max,
xmin = as.Date(-Inf), xmax = as.Date(Inf),
x = NULL, y = NULL, fill = color
)
) +
# use the hexidecimal colors from the dataset for the palette
scale_fill_identity() +
# format the x-axis for dates
scale_x_date(
name = NULL, date_labels = "%b %Y",
limits = c(ymd("2023-01-01"), ymd("2024-03-01"))
) +
# add text labels for each AQI category
geom_text(
data = aqi_levels,
aes(x = ymd("2024-02-28"), y = aqi_mid, label = level),
hjust = 1, size = 6, fontface = "bold", color = "white",
family = "Atkinson Hyperlegible"
) +
# plot the AQI in Syracuse
geom_line(linewidth = 1, alpha = 0.5) +
# human-readable labels
labs(
x = NULL, y = "AQI",
title = "Ozone and PM2.5 Daily AQI Values",
subtitle = "Syracuse, NY",
caption = "\nSource: EPA Daily Air Quality Tracker"
)Adjust plot theme components and use theme_minimal()
Your turn: Use theme_minimal() and get rid of the minor grid lines.
# draw the graph
syr_2023 |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
# shade in background with colors based on AQI guide
geom_rect(
data = aqi_levels,
aes(
ymin = aqi_min, ymax = aqi_max,
xmin = as.Date(-Inf), xmax = as.Date(Inf),
x = NULL, y = NULL, fill = color
)
) +
# use the hexidecimal colors from the dataset for the palette
scale_fill_identity() +
# format the x-axis for dates
scale_x_date(
name = NULL, date_labels = "%b %Y",
limits = c(ymd("2023-01-01"), ymd("2024-03-01"))
) +
# add text labels for each AQI category
geom_text(
data = aqi_levels,
aes(x = ymd("2024-02-28"), y = aqi_mid, label = level),
hjust = 1, size = 6, fontface = "bold", color = "white",
family = "Atkinson Hyperlegible"
) +
# plot the AQI in Syracuse
geom_line(linewidth = 1, alpha = 0.5) +
# human-readable labels
labs(
x = NULL, y = "AQI",
title = "Ozone and PM2.5 Daily AQI Values",
subtitle = "Syracuse, NY",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
# don't like the default theme
theme_minimal(base_size = 12, base_family = "Atkinson Hyperlegible") +
theme(
plot.title.position = "plot",
panel.grid.minor.y = element_blank(),
panel.grid.minor.x = element_blank()
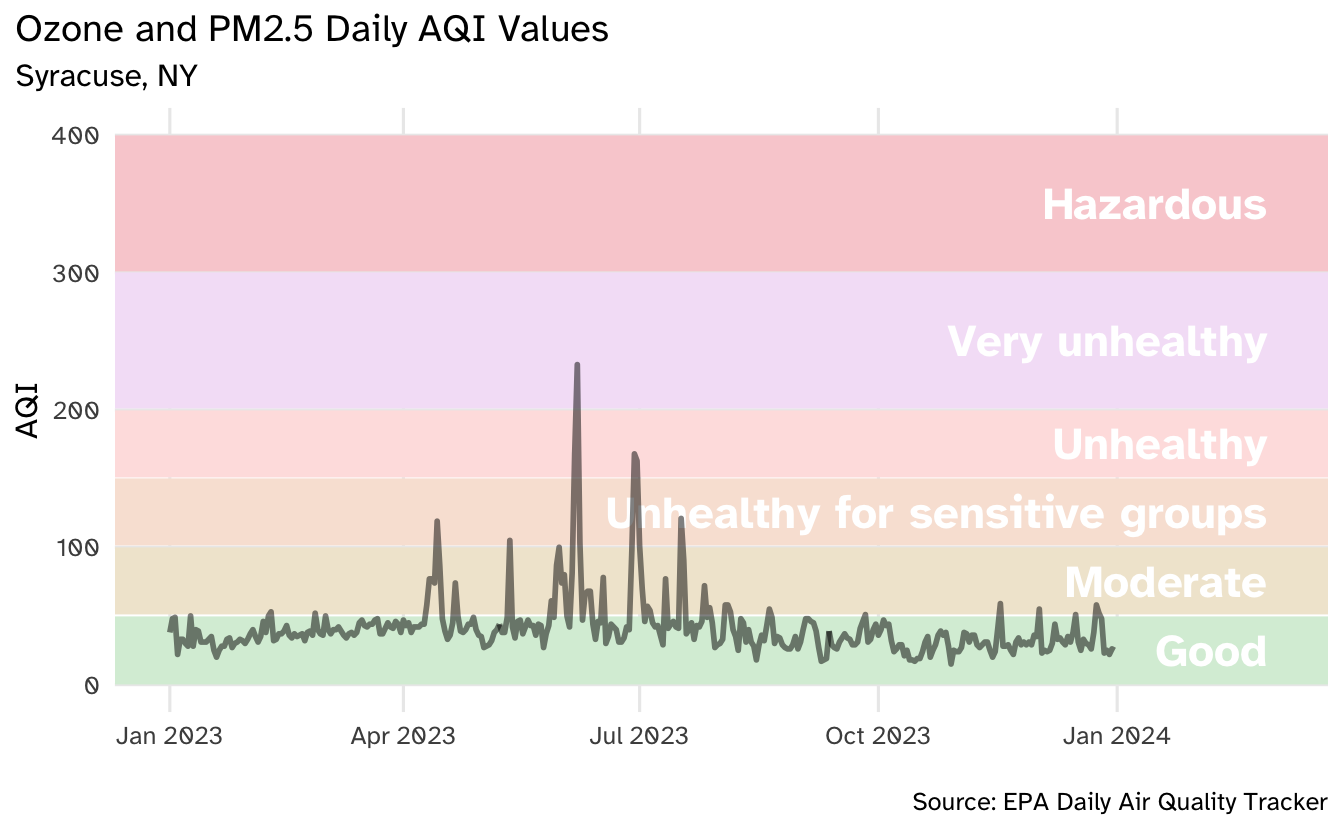
)Use fit-asp to ensure rendering is correct
Your turn: Render the plot with an aspect ratio of \(0.618\).
```{r}
#| label: plot-final
#| fig-asp: 0.618
aqi_levels <- aqi_levels |>
mutate(aqi_mid = ((aqi_min + aqi_max) / 2))
# draw the graph
syr_2023 |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
# shade in background with colors based on AQI guide
geom_rect(
data = aqi_levels,
aes(
ymin = aqi_min, ymax = aqi_max,
xmin = as.Date(-Inf), xmax = as.Date(Inf),
x = NULL, y = NULL, fill = color
)
) +
# use the hexidecimal colors from the dataset for the palette
scale_fill_identity() +
# format the x-axis for dates
scale_x_date(
name = NULL, date_labels = "%b %Y",
limits = c(ymd("2023-01-01"), ymd("2024-03-01"))
) +
# add text labels for each AQI category
geom_text(
data = aqi_levels,
aes(x = ymd("2024-02-28"), y = aqi_mid, label = level),
hjust = 1, size = 6, fontface = "bold", color = "white",
family = "Atkinson Hyperlegible"
) +
# plot the AQI in Syracuse
geom_line(linewidth = 1, alpha = 0.5) +
# human-readable labels
labs(
x = NULL, y = "AQI",
title = "Ozone and PM2.5 Daily AQI Values",
subtitle = "Syracuse, NY",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
# don't like the default theme
theme_minimal(base_size = 12, base_family = "Atkinson Hyperlegible") +
theme(
plot.title.position = "plot",
panel.grid.minor.y = element_blank(),
panel.grid.minor.x = element_blank()
)
```sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os macOS Ventura 13.5.2
system aarch64, darwin20
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz America/New_York
date 2024-03-21
pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.0)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.0)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.1)
colorspace * 2.1-0 2023-01-23 [1] CRAN (R 4.3.0)
crayon 1.5.2 2022-09-29 [1] CRAN (R 4.3.0)
digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.1)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.1)
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.1)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.1)
farver 2.1.1 2022-07-06 [1] CRAN (R 4.3.0)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.0)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.0)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.0)
ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.3.1)
glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.1)
gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.0)
here 1.0.1 2020-12-13 [1] CRAN (R 4.3.0)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.0)
htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.1)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.1)
janitor * 2.2.0 2023-02-02 [1] CRAN (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.1)
knitr 1.45 2023-10-30 [1] CRAN (R 4.3.1)
labeling 0.4.3 2023-08-29 [1] CRAN (R 4.3.0)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.1)
lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.1)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.0)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.0)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.0)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.0)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.1)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.1)
rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.1)
rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.3.1)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.0)
scales * 1.2.1 2024-01-18 [1] Github (r-lib/scales@c8eb772)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.0)
snakecase 0.11.1 2023-08-27 [1] CRAN (R 4.3.0)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.1)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.1)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.0)
tidyr * 1.3.0 2023-01-24 [1] CRAN (R 4.3.0)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.0)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.0)
timechange 0.2.0 2023-01-11 [1] CRAN (R 4.3.0)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.0)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.1)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.1)
vroom 1.6.5 2023-12-05 [1] CRAN (R 4.3.1)
withr 2.5.2 2023-10-30 [1] CRAN (R 4.3.1)
xfun 0.41 2023-11-01 [1] CRAN (R 4.3.1)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.1)
[1] /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/library
──────────────────────────────────────────────────────────────────────────────