Explainable machine learning models
Lecture 25
Dr. Benjamin Soltoff
Cornell University
INFO 3312/5312 - Spring 2024
May 4, 2023
Announcements
Announcements
- Presentations tomorrow
- Course evaluations – 0.5% extra credit towards your final grade if you complete your course evaluation by May 12
Setup
Packages + figures
YAML options
Interpretability and explainability
Interpretation
- Interpretability is the degree to which a human can understand the cause of a decision
- Interpretability is the degree to which a human can consistently predict the model’s result
- How does this model work?
Explanation
Answer to the “why” question
- Why did the government collapse?
- Why was my loan rejected?
- Focuses on a single prediction
What is a good explanation?
- Contrastive: why was this prediction made instead of another prediction?
- Selected: Focuses on just a handful of reasons, even if the problem is more complex
- Social: Needs to be understandable by your audience
- Truthful: Explanation should predict the event as truthfully as possible
- Generalizable: Explanation could apply to many predictions
Global vs. local methods
- Interpretation \(\leadsto\) global methods
- Explanation \(\leadsto\) local methods
White-box model
Models that lend themselves naturally to interpretation
- Linear regression
- Logistic regression
- Generalized linear model
- Decision tree
Black-box model

Black-box model
- Random forests
- Boosted trees
- Neural networks
- Deep learning

Predicting student debt
- College Scorecard
- rscorecard
rcis::scorecard
Fitted models
library(tidyverse)
library(tidymodels)
library(rcis)
library(here)
# get scorecard dataset
data("scorecard")
scorecard <- scorecard |>
# remove ID columns - causing issues when interpreting/explaining
select(-unitid, -name) |>
# convert factor to character columns
mutate(across(.cols = where(is.factor), .f = as.character)) |>
# remove any rows with missing values - just makes life easier for explanation methods
drop_na()
# split into training and testing
set.seed(123)
scorecard_split <- initial_split(data = scorecard, prop = .75, strata = debt)
scorecard_train <- training(scorecard_split)
scorecard_test <- testing(scorecard_split)
scorecard_folds <- vfold_cv(data = scorecard_train, v = 10)
# basic feature engineering recipe
scorecard_rec <- recipe(debt ~ ., data = scorecard_train) |>
# catch all category for missing state values
step_novel(state) |>
# use median imputation for numeric predictors
step_impute_median(all_numeric_predictors()) |>
# use modal imputation for nominal predictors
step_impute_mode(all_nominal_predictors()) |>
# remove rows with missing values for
# outcomes - glmnet won't work if any of this column is NA
step_naomit(all_outcomes())
# generate random forest model
rf_mod <- rand_forest() |>
set_engine("ranger") |>
set_mode("regression")
# combine recipe with model
rf_wf <- workflow() |>
add_recipe(scorecard_rec) |>
add_model(rf_mod)
# fit using training set
set.seed(123)
rf_wf <- fit(
rf_wf,
data = scorecard_train
)
# fit penalized regression model
## recipe
glmnet_recipe <- scorecard_rec |>
step_dummy(all_nominal_predictors()) |>
step_zv(all_predictors()) |>
step_normalize(all_numeric_predictors())
## model specification
glmnet_spec <- linear_reg(penalty = tune(), mixture = tune()) |>
set_mode("regression") |>
set_engine("glmnet")
## workflow
glmnet_workflow <- workflow() |>
add_recipe(glmnet_recipe) |>
add_model(glmnet_spec)
## tuning grid
glmnet_grid <- expand_grid(
penalty = 10^seq(-6, -1, length.out = 20),
mixture = c(0.05, 0.2, 0.4, 0.6, 0.8, 1)
)
## hyperparameter tuning
glmnet_tune <- tune_grid(
glmnet_workflow,
resamples = scorecard_folds,
grid = glmnet_grid
)
# select best model
glmnet_best <- select_best(glmnet_tune, metric = "rmse")
glmnet_wf <- finalize_workflow(glmnet_workflow, glmnet_best) |>
last_fit(scorecard_split) |>
extract_workflow()
# nearest neighbors model
## use glmnet recipe
kknn_spec <- nearest_neighbor(neighbors = 10) |>
set_mode("regression") |>
set_engine("kknn")
kknn_workflow <-
workflow() |>
add_recipe(glmnet_recipe) |>
add_model(kknn_spec)
## fit using training set
set.seed(123)
kknn_wf <- fit(
kknn_workflow,
data = scorecard_train
)
# save all required objects to a .Rdata file
save(scorecard_train, scorecard_test, rf_wf, glmnet_wf, kknn_wf,
file = here("slides/data/scorecard-models.RData"))Predicting student debt
Rows: 1,721
Columns: 14
$ unitid <dbl> 100654, 100663, 100706, 100724, 100751, 100830, 100858, 1009…
$ name <chr> "Alabama A & M University", "University of Alabama at Birmin…
$ state <chr> "AL", "AL", "AL", "AL", "AL", "AL", "AL", "AL", "AL", "AL", …
$ type <fct> "Public", "Public", "Public", "Public", "Public", "Public", …
$ admrate <dbl> 0.8965, 0.8060, 0.7711, 0.9888, 0.8039, 0.9555, 0.8507, 0.60…
$ satavg <dbl> 959, 1245, 1300, 938, 1262, 1061, 1302, 1202, 1068, NA, 1101…
$ cost <dbl> 23445, 25542, 24861, 21892, 30016, 20225, 32196, 32514, 3483…
$ netcost <dbl> 15529, 16530, 17208, 19534, 20917, 13678, 24018, 19808, 2050…
$ avgfacsal <dbl> 68391, 102420, 87273, 64746, 93141, 69561, 96498, 62649, 533…
$ pctpell <dbl> 0.7095, 0.3397, 0.2403, 0.7368, 0.1718, 0.4654, 0.1343, 0.22…
$ comprate <dbl> 0.2866, 0.6117, 0.5714, 0.3177, 0.7214, 0.3040, 0.7870, 0.70…
$ firstgen <dbl> 0.3658281, 0.3412237, 0.3101322, 0.3434343, 0.2257127, 0.381…
$ debt <dbl> 15250, 15085, 14000, 17500, 17671, 12000, 17500, 16000, 1425…
$ locale <fct> City, City, City, City, City, City, City, City, City, Suburb…Evaluation performance

Global interpretation methods
Permutation-based feature importance
- Calculate the increase in the model’s prediction error after permuting the feature
- Randomly shuffle the feature’s values across observations
- Important feature
- Unimportant feature
For any given loss function do
1: compute loss function for original model
2: for variable i in {1,...,p} do
| randomize values
| apply given ML model
| estimate loss function
| compute feature importance (permuted loss / original loss)
end
3. Sort variables by descending feature importance Random forest feature importance
Code
explainer_glmnet <- explain_tidymodels(
model = glmnet_wf,
data = scorecard_train |> select(-debt),
y = scorecard_train$debt,
label = "penalized regression",
verbose = FALSE
)
explainer_rf <- explain_tidymodels(
model = rf_wf,
data = scorecard_train |> select(-debt),
y = scorecard_train$debt,
label = "random forest",
verbose = FALSE
)
explainer_kknn <- explain_tidymodels(
model = kknn_wf,
data = scorecard_train |> select(-debt),
y = scorecard_train$debt,
label = "k nearest neighbors",
verbose = FALSE
)
Number of observations permuted
model_parts(explainer_rf, N = 100) |>
plot() +
labs(
title = "N = 100",
subtitle = NULL
)
model_parts(explainer_rf, N = 200) |>
plot() +
labs(
title = "N = 200",
subtitle = NULL
)
model_parts(explainer_rf, N = NULL) |>
plot() +
labs(
title = "N = NULL",
subtitle = NULL
)Code

Code

Code

Compare all models
Code
# plot variable importance
# source: https://www.tmwr.org/explain.html#global-explanations
ggplot_imp <- function(...) {
obj <- list(...)
metric_name <- attr(obj[[1]], "loss_name")
metric_lab <- paste(
metric_name,
"after permutations\n(higher indicates more important)"
)
full_vip <- bind_rows(obj) |>
filter(variable != "_baseline_")
perm_vals <- full_vip |>
filter(variable == "_full_model_") |>
group_by(label) |>
summarise(dropout_loss = mean(dropout_loss))
p <- full_vip |>
filter(variable != "_full_model_") |>
mutate(variable = fct_reorder(variable, dropout_loss)) |>
ggplot(aes(dropout_loss, variable))
if (length(obj) > 1) {
p <- p +
facet_wrap(vars(label)) +
geom_vline(
data = perm_vals, aes(xintercept = dropout_loss, color = label),
size = 1.4, lty = 2, alpha = 0.7
) +
geom_boxplot(aes(color = label, fill = label), alpha = 0.2)
} else {
p <- p +
geom_vline(
data = perm_vals, aes(xintercept = dropout_loss),
size = 1.4, lty = 2, alpha = 0.7
) +
geom_boxplot(fill = "#91CBD765", alpha = 0.4)
}
p +
theme(legend.position = "none") +
labs(
x = metric_lab,
y = NULL, fill = NULL, color = NULL
)
}
vip_rf <- model_parts(explainer_rf, N = NULL)
vip_glmnet <- model_parts(explainer_glmnet, N = NULL)
vip_kknn <- model_parts(explainer_kknn, N = NULL)
ggplot_imp(vip_rf, vip_glmnet, vip_kknn)
Individual conditional expectation
- Ceteris peribus - “other things held constant”
- Marginal effect a feature has on the predictor
- Plot one line per observation that shows how the observation’s prediction changes when a feature changes
- Partial dependence plot is average of all ICEs
For a selected predictor (x)
1. Determine grid space of j evenly spaced values across distribution of x
2: for value i in {1,...,j} of grid space do
| set x to i for all observations
| apply given ML model
| estimate predicted value
| if PDP: average predicted values across all observations
endNet cost (PDP)

Net cost (PDP + ICE)

Net cost (PDP + ICE) – all models
model_profile(explainer_rf, variables = "netcost", N = NULL) |> plot(geom = "profiles")
model_profile(explainer_glmnet, variables = "netcost", N = NULL) |> plot(geom = "profiles")
model_profile(explainer_kknn, variables = "netcost", N = NULL) |> plot(geom = "profiles")Code

Code

Code

Type (PDP)

State (PDP)
Code
# PDP for state
## hard to read
pdp_state_kknn <- model_profile(explainer_kknn, variables = "state", N = NULL)
## manually construct and reorder states
## extract aggregated profiles
pdp_state_kknn$agr_profiles |>
# convert to tibble
as_tibble() |>
mutate(`_x_` = fct_reorder(.f = `_x_`, .x = `_yhat_`)) |>
ggplot(mapping = aes(x = `_yhat_`, y = `_x_`, fill = `_yhat_`)) +
geom_col() +
scale_x_continuous(labels = scales::dollar) +
scale_fill_viridis_c(guide = "none") +
labs(
title = "Partial dependence plot for state",
subtitle = "Created for the k nearest neighbors model",
x = "Average prediction",
y = NULL
) +
theme_minimal(base_size = 9)
Local explanatory methods
Code
data("scorecard")
cornell <- filter(.data = scorecard, name == "Cornell University") |>
select(-unitid, -name)
ith_coll <- filter(.data = scorecard, name == "Ithaca College") |>
select(-unitid, -name)
both <- bind_rows(cornell, ith_coll)
# set row names for LIME later
rownames(both) <- c("Cornell University", "Ithaca College")Cornell University
Rows: 1
Columns: 12
$ state <chr> "NY"
$ type <fct> "Private, nonprofit"
$ admrate <dbl> 0.1071
$ satavg <dbl> 1480
$ cost <dbl> 75204
$ netcost <dbl> 37042
$ avgfacsal <dbl> 137169
$ pctpell <dbl> 0.1622
$ comprate <dbl> 0.9536
$ firstgen <dbl> 0.154164
$ debt <dbl> 13108
$ locale <fct> CityIthaca College
Rows: 1
Columns: 12
$ state <chr> "NY"
$ type <fct> "Private, nonprofit"
$ admrate <dbl> 0.7568
$ satavg <dbl> NA
$ cost <dbl> 64006
$ netcost <dbl> 34554
$ avgfacsal <dbl> 82620
$ pctpell <dbl> 0.1936
$ comprate <dbl> 0.7752
$ firstgen <dbl> 0.1375752
$ debt <dbl> 19500
$ locale <fct> SuburbBreakdown methods
- How contributions attributed to individual features change the mean model’s prediction for a particular observation
- Sequentially fix the value of individual features and examine the change in
Breakdown of random forest
bd1_rf_distr <- predict_parts(
explainer = explainer_rf,
new_observation = cornell,
type = "break_down",
order = NULL,
keep_distributions = TRUE
)
plot(bd1_rf_distr, plot_distributions = TRUE)
plot(bd1_rf_distr)Code

Code

Breakdown of random forest
bd2_rf_distr <- predict_parts(
explainer = explainer_rf,
new_observation = cornell,
type = "break_down",
order = names(cornell),
keep_distributions = TRUE
)
plot(bd2_rf_distr, plot_distributions = TRUE)
plot(bd2_rf_distr)Code

Code

Breakdown of random forest
rsample <- map(1:6, function(i) {
new_order <- sample(1:12)
bd <- predict_parts(explainer_rf, cornell, order = new_order, type = "break_down")
bd$variable <- as.character(bd$variable)
bd$label <- paste("random order no.", i)
plot(bd) +
theme_minimal(base_size = 11) +
theme(legend.position = "none")
})
map(.x = rsample, .f = print)Code






[[1]]
[[2]]
[[3]]
[[4]]
[[5]]
[[6]]
Shapley Additive Explanations (SHAP)
- Average contributions of features are computed under different coalitions of feature orderings
- Randomly permute feature order using \(B\) combinations
- Average across individual breakdowns to calculate feature contribution to individual prediction
Shapley Additive Explanations (SHAP)
# explain cornell with rf and kknn models
shap_cornell_rf <- predict_parts(
explainer = explainer_rf,
new_observation = cornell,
type = "shap"
)
shap_cornell_kknn <- predict_parts(
explainer = explainer_kknn,
new_observation = cornell,
type = "shap"
)
plot(shap_cornell_rf)
plot(shap_cornell_kknn)Code


Shapley Additive Explanations (SHAP)
# explain cornell with rf model
shap_ith_coll_rf <- predict_parts(
explainer = explainer_rf,
new_observation = ith_coll,
type = "shap"
)
plot(shap_cornell_rf) +
ggtitle("Cornell University")
plot(shap_ith_coll_rf) +
ggtitle("Ithaca College")Code


LIME
Local interpretable model-agnostic explanations
- Global \(\rightarrow\) local
- Interpretable model used to explain individual predictions of a black box model
- Assumes every complex model is linear on a local scale
- Simple model explains the predictions of the complex model locally
- Local fidelity
- Does not require global fidelity
- Works on tabular, text, and image data
LIME
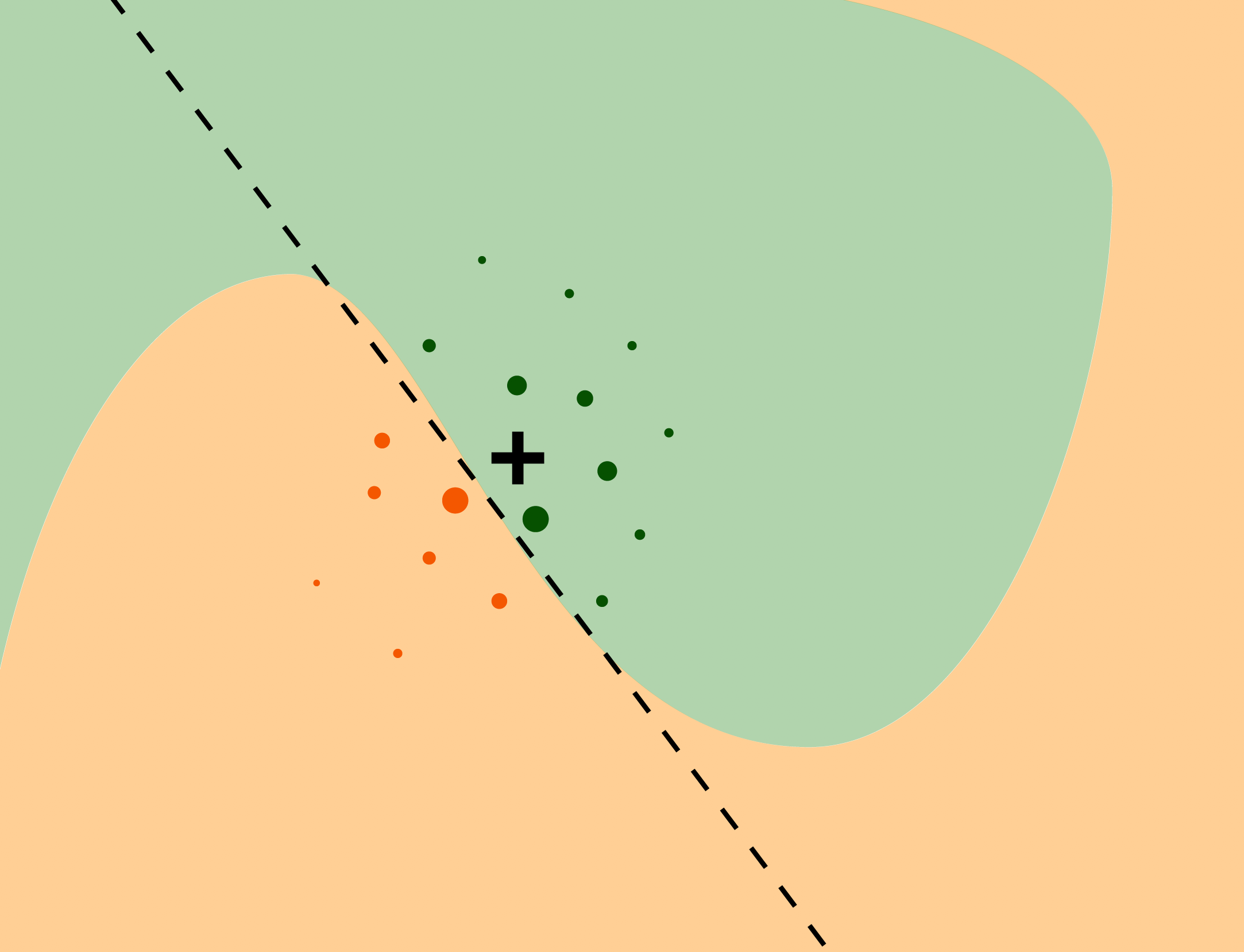
Source: Explanatory Model Analysis
LIME
- For each prediction to explain, permute the observation \(n\) times
- Let the complex model predict the outcome of all permuted observations
- Calculate the distance from all permutations to the original observation
- Convert the distance to a similarity score
- Select \(m\) features best describing the complex model outcome from the permuted data
- Fit a simple model to the permuted data, explaining the complex model outcome with the \(m\) features from the permuted data weighted by its similarity to the original observation
- Extract the feature weights from the simple model and use these as explanations for the complex models local behavior
\(10\) nearest neighbors
Code
# prepare the recipe
prepped_rec_kknn <- extract_recipe(kknn_wf)
# write a function to bake the observation
bake_kknn <- function(x) {
bake(
prepped_rec_kknn,
new_data = x
)
}
# create explainer object
lime_explainer_kknn <- lime(
x = scorecard_train,
model = extract_fit_parsnip(kknn_wf),
preprocess = bake_kknn
)
# top 5 features
explanation_kknn <- explain(
x = both,
explainer = lime_explainer_kknn,
n_features = 10
)
plot_features(explanation_kknn) +
theme_minimal(base_size = 12) +
theme(legend.position = "bottom")
Random forest
Code
# prepare the recipe
prepped_rec_rf <- extract_recipe(rf_wf)
# write a function to bake the observation
bake_rf <- function(x) {
bake(
prepped_rec_rf,
new_data = x
)
}
# create explainer object
lime_explainer_rf <- lime(
x = scorecard_train,
model = extract_fit_parsnip(rf_wf),
preprocess = bake_rf
)
# top 5 features
explanation_rf <- explain(
x = both,
explainer = lime_explainer_rf,
n_features = 10
)
plot_features(explanation_rf) +
theme_minimal(base_size = 12) +
theme(legend.position = "bottom")
Additional resources
Underlying methods