Visualizing time series data
Lecture 15
Dr. Benjamin Soltoff
Cornell University
INFO 3312/5312 - Spring 2026
March 19, 2024
Announcements
Announcements
- Nothing
Visualization critique
Blame Canada! Blame Canada!
Working with dates
Air Quality Index
The AQI is the Environmental Protection Agency’s index for reporting air quality
Higher values of AQI indicate worse air quality

AQI levels
The previous graphic in tibble form, to be used later…
AQI data
Source: EPA’s Daily Air Quality Tracker
2023 AQI (Ozone and PM2.5 combined) for Syracuse, NY
2023 Syracuse
- Load data
Clean variable names
First look
This plot looks quite bizarre. What might be going on?
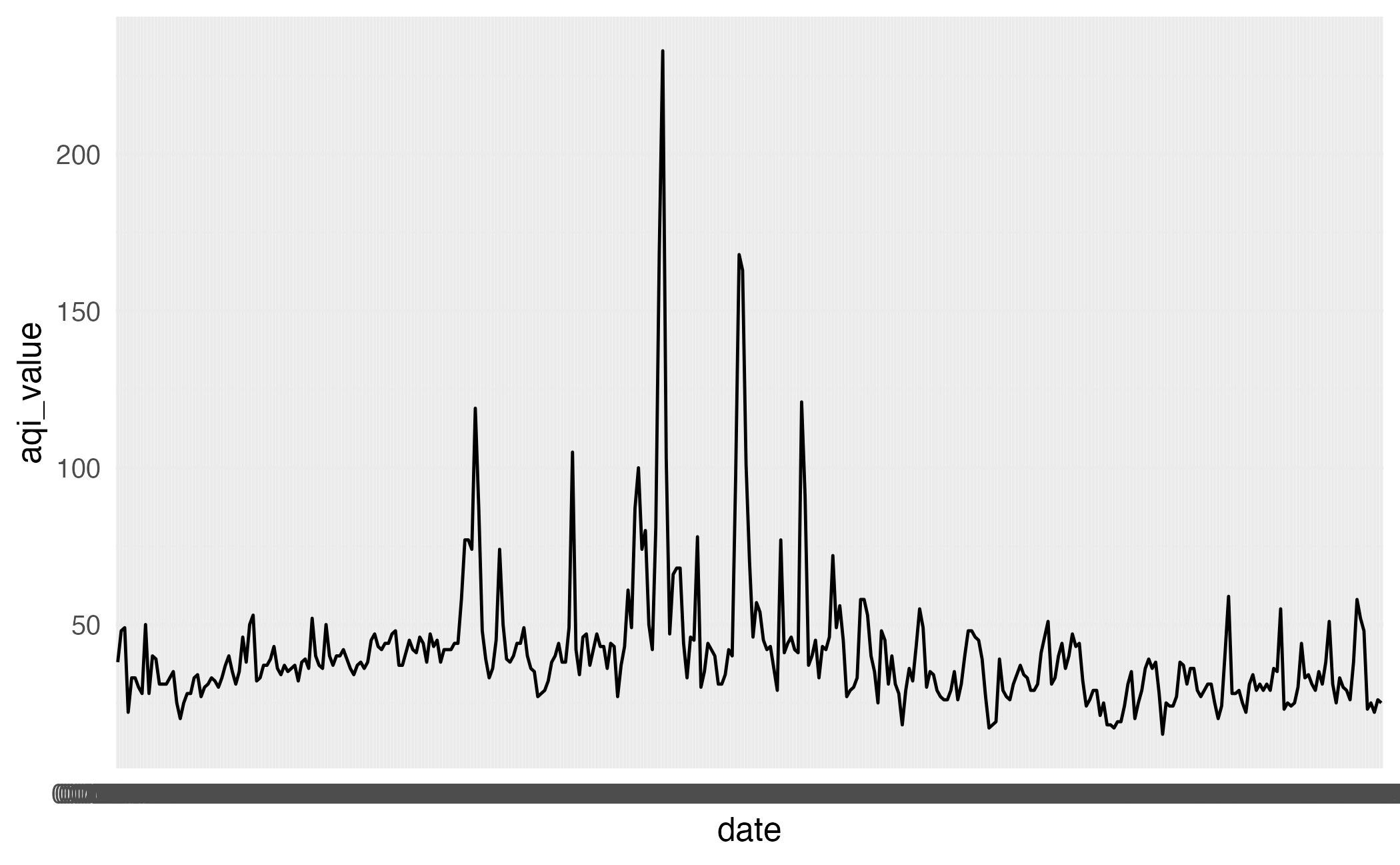
Peek at data
# A tibble: 365 × 4
date aqi_value site_name site_id
<chr> <dbl> <chr> <chr>
1 01/01/2023 38 EAST SYRACUSE 36-067-1015
2 01/02/2023 48 EAST SYRACUSE 36-067-1015
3 01/03/2023 49 EAST SYRACUSE 36-067-1015
4 01/04/2023 22 EAST SYRACUSE 36-067-1015
5 01/05/2023 33 EAST SYRACUSE 36-067-1015
6 01/06/2023 33 EAST SYRACUSE 36-067-1015
7 01/07/2023 30 EAST SYRACUSE 36-067-1015
8 01/08/2023 28 EAST SYRACUSE 36-067-1015
9 01/09/2023 50 EAST SYRACUSE 36-067-1015
10 01/10/2023 28 FULTON 36-075-0003
# ℹ 355 more rowsTransforming date
Using lubridate::mdy():
# A tibble: 365 × 11
date aqi_value main_pollutant site_name site_id source x20_year_high_2000_2019
<date> <dbl> <chr> <chr> <chr> <chr> <dbl>
1 2023-01-01 38 PM2.5 EAST SYRACUSE 36-067-1015 AQS 54
2 2023-01-02 48 PM2.5 EAST SYRACUSE 36-067-1015 AQS 43
3 2023-01-03 49 PM2.5 EAST SYRACUSE 36-067-1015 AQS 35
4 2023-01-04 22 PM2.5 EAST SYRACUSE 36-067-1015 AQS 52
5 2023-01-05 33 PM2.5 EAST SYRACUSE 36-067-1015 AQS 64
6 2023-01-06 33 PM2.5 EAST SYRACUSE 36-067-1015 AQS 67
7 2023-01-07 30 PM2.5 EAST SYRACUSE 36-067-1015 AQS 77
8 2023-01-08 28 PM2.5 EAST SYRACUSE 36-067-1015 AQS 38
9 2023-01-09 50 PM2.5 EAST SYRACUSE 36-067-1015 AQS 55
10 2023-01-10 28 Ozone FULTON 36-075-0003 AQS 65
# ℹ 355 more rows
# ℹ 4 more variables: x20_year_low_2000_2019 <dbl>, x5_year_average_2015_2019 <dbl>,
# date_of_20_year_high <chr>, date_of_20_year_low <chr>Data cleaning
# A tibble: 365 × 11
date aqi_value main_pollutant site_name site_id source x20_year_high_2000_2019
<date> <dbl> <chr> <chr> <chr> <chr> <dbl>
1 2023-01-01 38 PM2.5 EAST SYRACUSE 36-067-1015 AQS 54
2 2023-01-02 48 PM2.5 EAST SYRACUSE 36-067-1015 AQS 43
3 2023-01-03 49 PM2.5 EAST SYRACUSE 36-067-1015 AQS 35
4 2023-01-04 22 PM2.5 EAST SYRACUSE 36-067-1015 AQS 52
5 2023-01-05 33 PM2.5 EAST SYRACUSE 36-067-1015 AQS 64
6 2023-01-06 33 PM2.5 EAST SYRACUSE 36-067-1015 AQS 67
7 2023-01-07 30 PM2.5 EAST SYRACUSE 36-067-1015 AQS 77
8 2023-01-08 28 PM2.5 EAST SYRACUSE 36-067-1015 AQS 38
9 2023-01-09 50 PM2.5 EAST SYRACUSE 36-067-1015 AQS 55
10 2023-01-10 28 Ozone FULTON 36-075-0003 AQS 65
# ℹ 355 more rows
# ℹ 4 more variables: x20_year_low_2000_2019 <dbl>, x5_year_average_2015_2019 <dbl>,
# date_of_20_year_high <chr>, date_of_20_year_low <chr>Another look
How would you improve this visualization?
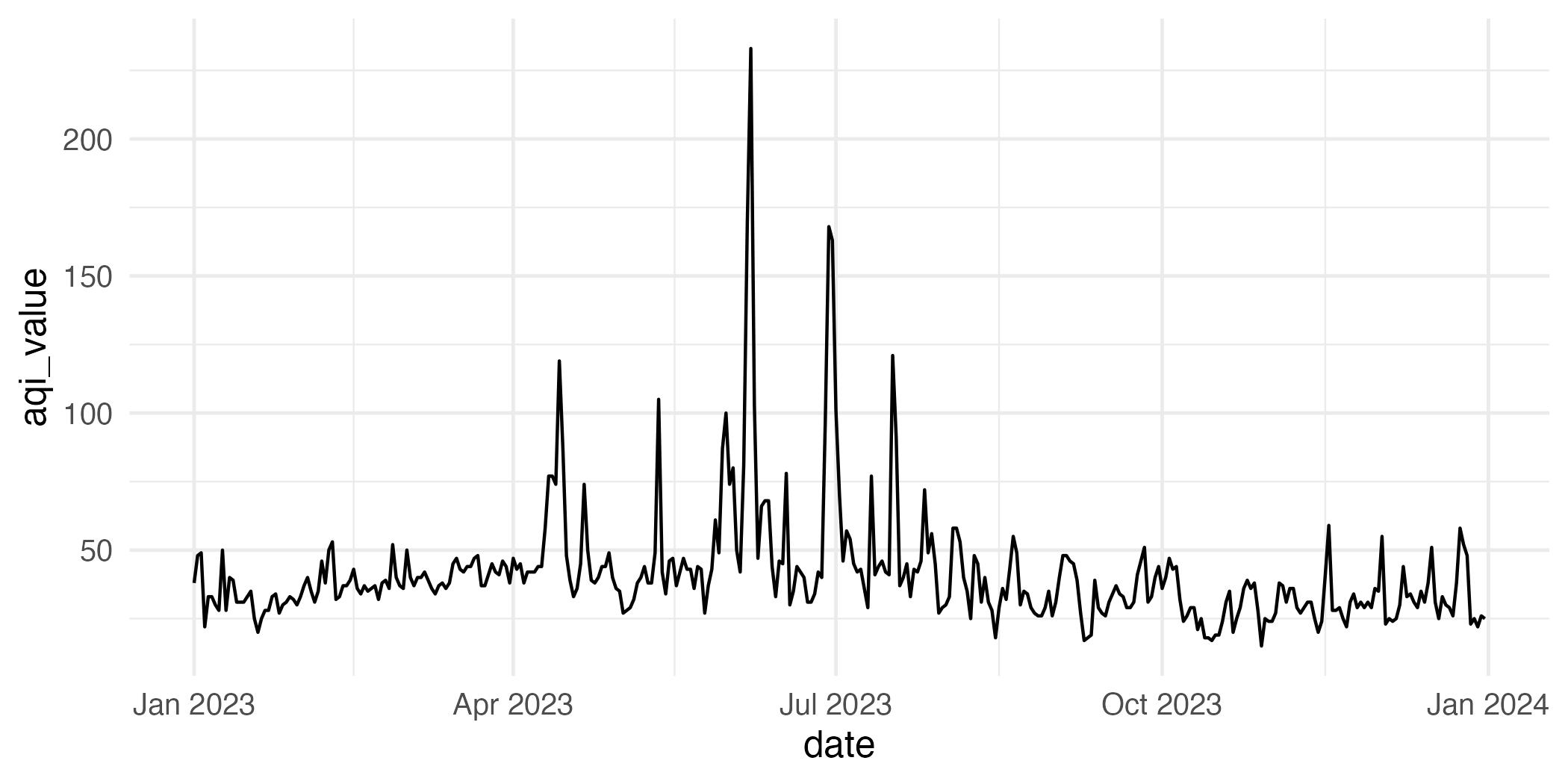
Application exercise
Visualizing Syracuse AQI
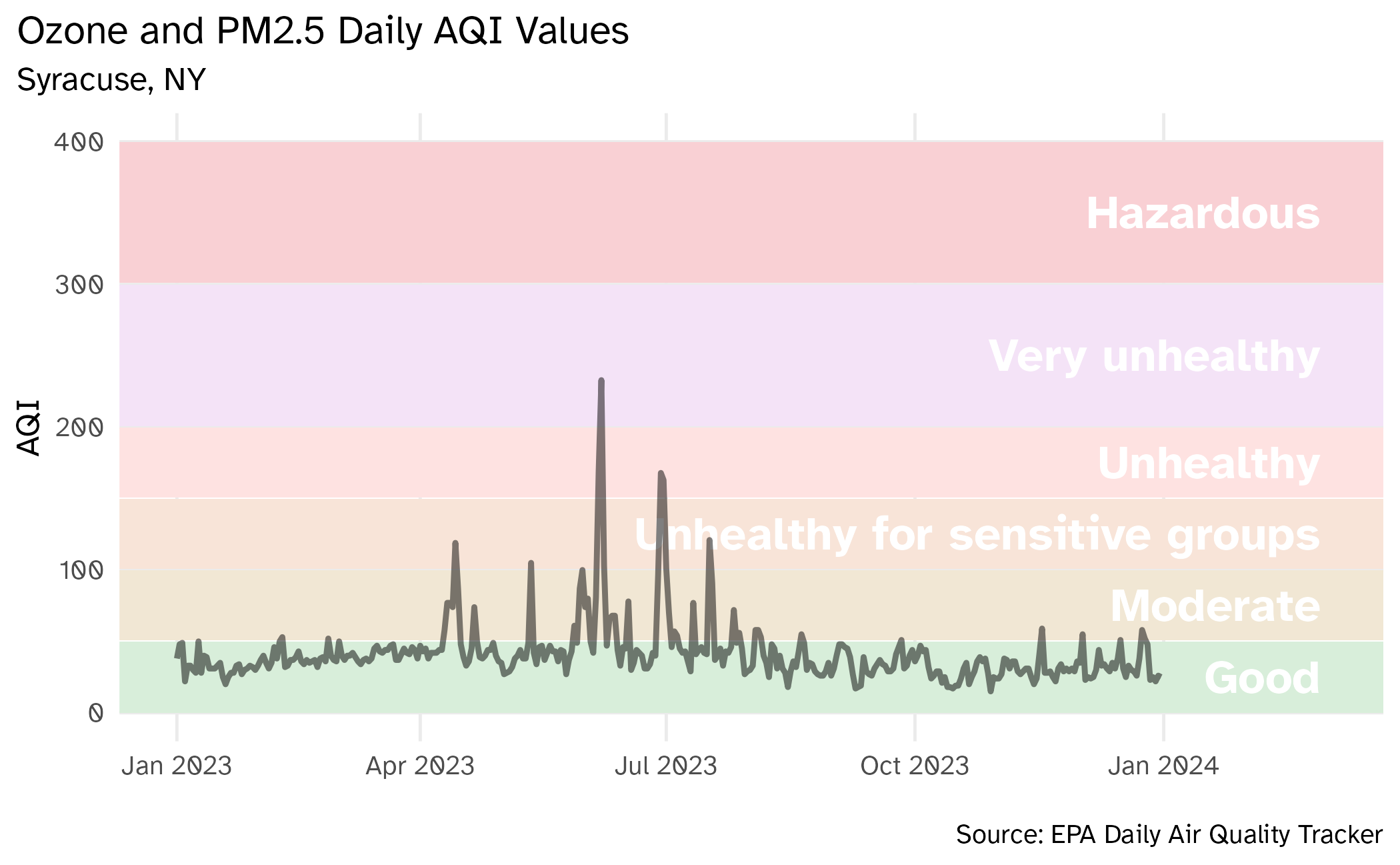
ae-12
- Go to the course GitHub org and find your
ae-12(repo name will be suffixed with your NetID). - Clone the repo in RStudio Workbench, open the Quarto document in the repo, and follow along and complete the exercises.
- Render, commit, and push your edits by the AE deadline – end of tomorrow.
Livecoding
Reveal below for code developed during live coding session.
Visualizing Syracuse AQI (take 2)
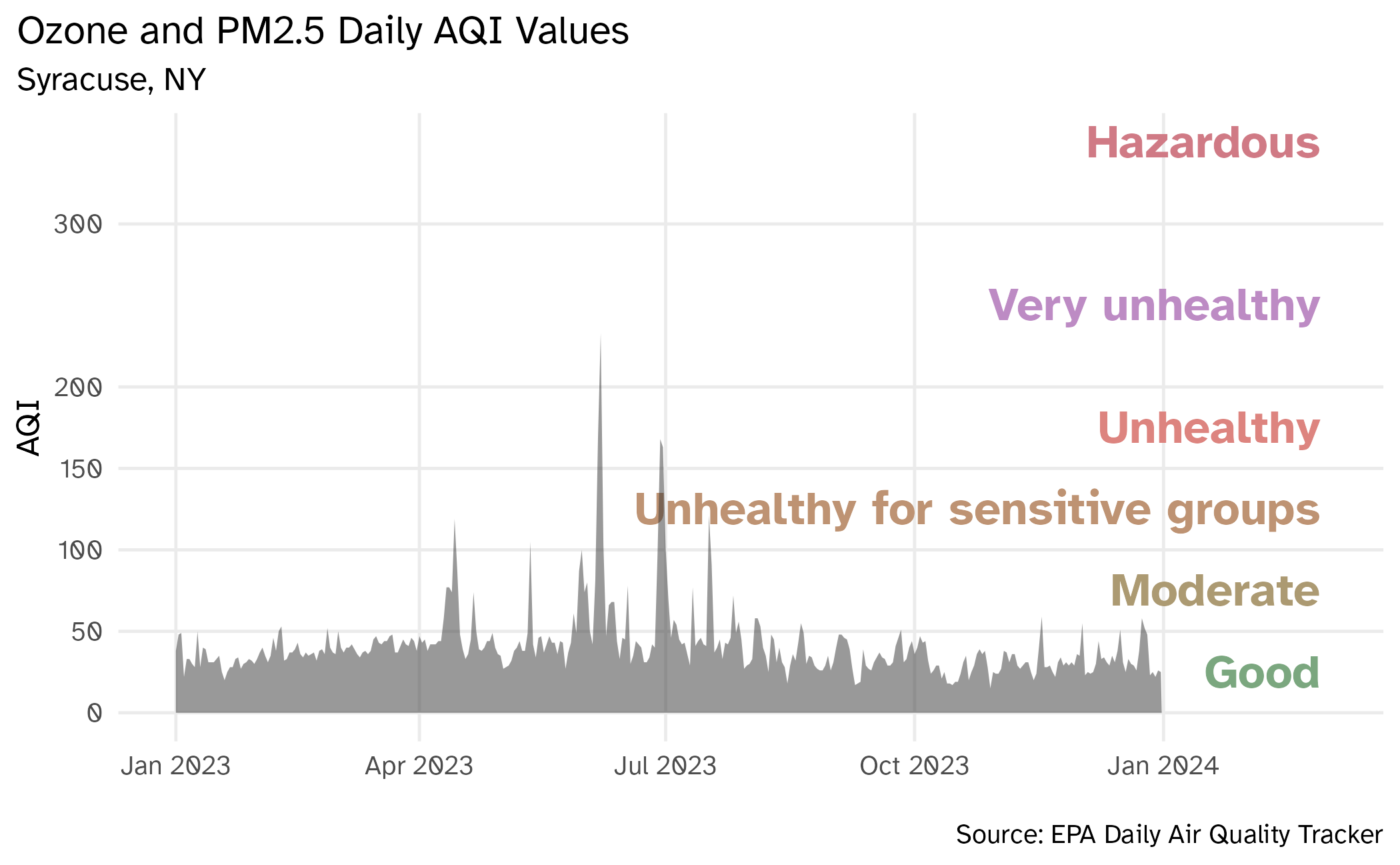
aqi_levels <- aqi_levels |>
mutate(aqi_mid = ((aqi_min + aqi_max) / 2))
# draw the graph
syr_2023 |>
# remove rows with missing AQIs
drop_na(aqi_value) |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
# add breaks and labels for AQI levels
scale_y_continuous(breaks = c(0, 50, 100, 150, 200, 300, 400)) +
geom_text(
data = aqi_levels,
aes(
x = ymd("2024-02-28"), y = aqi_mid,
label = level, color = darken(color, 0.3)
),
hjust = 1, size = 6,
family = "Atkinson Hyperlegible", fontface = "bold"
) +
# use the hexidecimal colors from the dataset for the palette
scale_color_identity() +
# format the x-axis for dates
scale_x_date(
name = NULL, date_labels = "%b %Y",
limits = c(ymd("2023-01-01"), ymd("2024-03-01"))
) +
# plot the AQI in Syracuse
geom_area(linewidth = 1, alpha = 0.5) +
# human-readable labels
labs(
x = NULL, y = "AQI",
title = "Ozone and PM2.5 Daily AQI Values",
subtitle = "Syracuse, NY",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
# don't like the default theme
theme_minimal(base_size = 12, base_family = "Atkinson Hyperlegible") +
theme(
plot.title.position = "plot",
panel.grid.minor.y = element_blank(),
panel.grid.minor.x = element_blank()
)Wrap-up
Wrap-up
- Ensure dates/times are structured correctly using {lubridate}
- Clearly depict the temporal flow of time in the chart
- More advanced methods - learn time series regression (STSCI 4550/5550)
