Deep dive: coordinates + facets
Lecture 6
Cornell University
INFO 3312/5312 - Spring 2026
February 5, 2026
Announcements
Announcements
- Homework 03
- Project 01 begin tomorrow – look for team assignments in Canvas
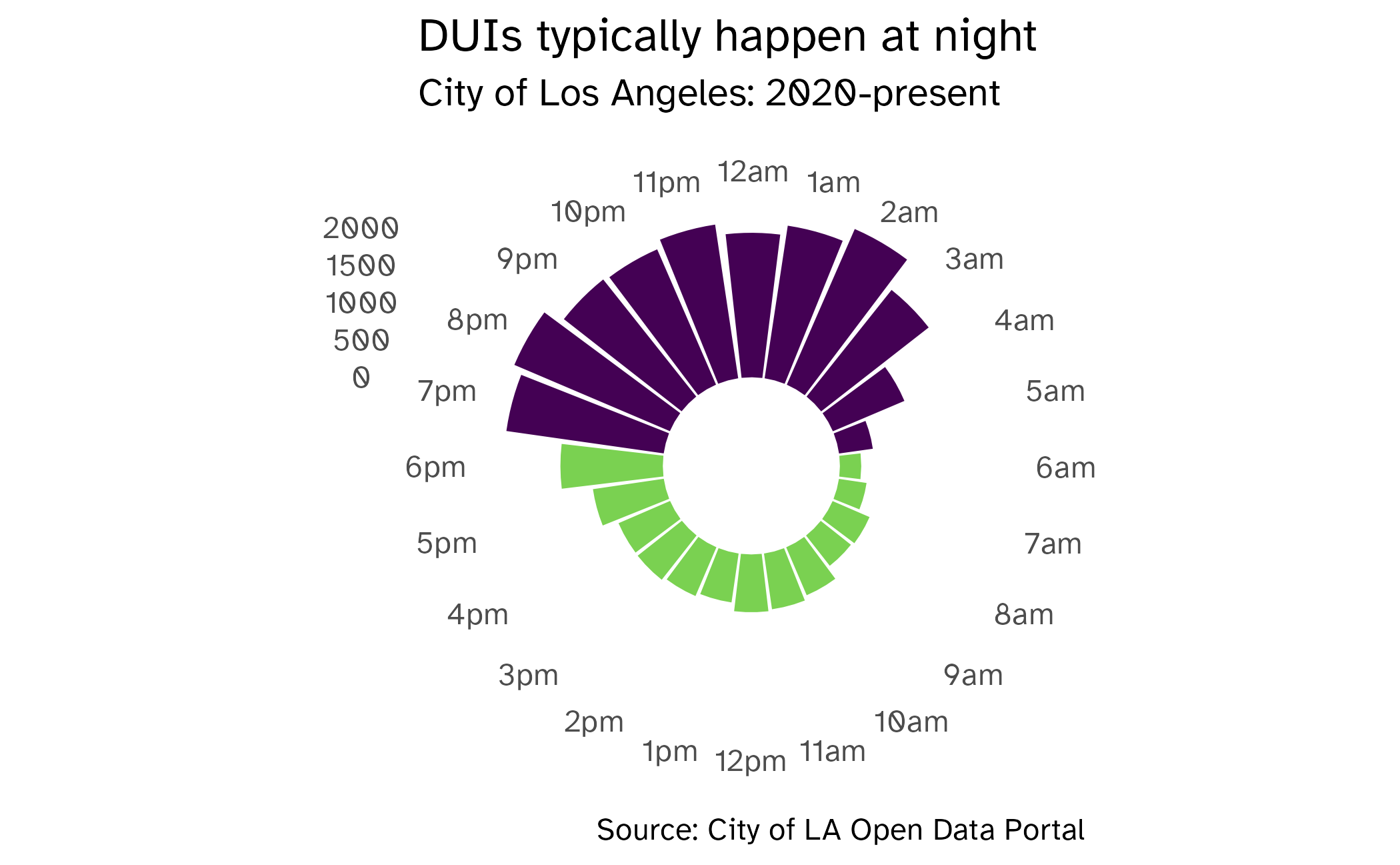
Where did the school buses go?
Source: Andrew Van Dam
Learning objectives
- Define coordinate systems
- Identify methods for implementing non-Cartesian coordinate systems with {ggplot2}
- Implement waffle charts
- Utilize facets for small multiple plots
Coordinate systems
Coordinate systems: purpose
- Combine the two position aesthetics (
xandy) to produce a two-dimension position on the plot- Linear coordinate system: horizontal and vertical coordinates
- Polar coordinate system: angle and radius
- Maps: latitude and longitude
- Draw axes and panel backgrounds in coordination with the coordinate systems
Linear coordinate systems
Preserve the shape of geoms
coord_cartesian(): the default Cartesian coordinate system, where the 2D position of an element is given by the combination of thexandypositions.coord_flip(): Cartesian coordinate system withxandyaxes flipped.- Just swap the
xandyaesthetics instead
- Just swap the
coord_fixed(): Cartesian coordinate system with a fixed aspect ratio.- Use
coord_cartesian(ratio = X)to set a specific aspect ratio.
- Use
Non-linear coordinate systems
Can change the shapes – a straight line may no longer be straight. The closest distance between two points may no longer be a straight line.
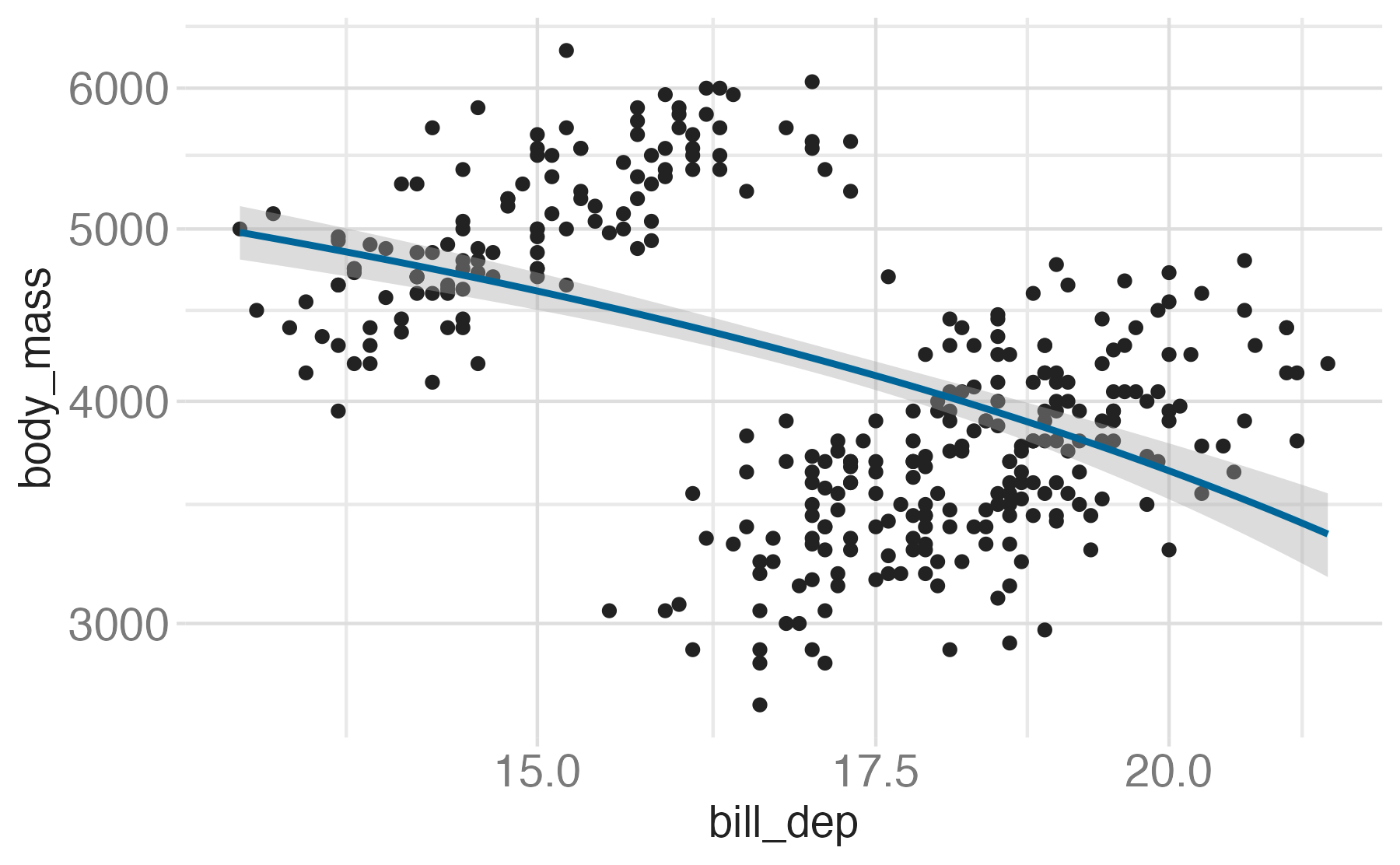
coord_transform(): Apply arbitrary transformations to x and y positions, after the data has been processed by the statcoord_radial(): Polar coordinatescoord_sf(): Map projections
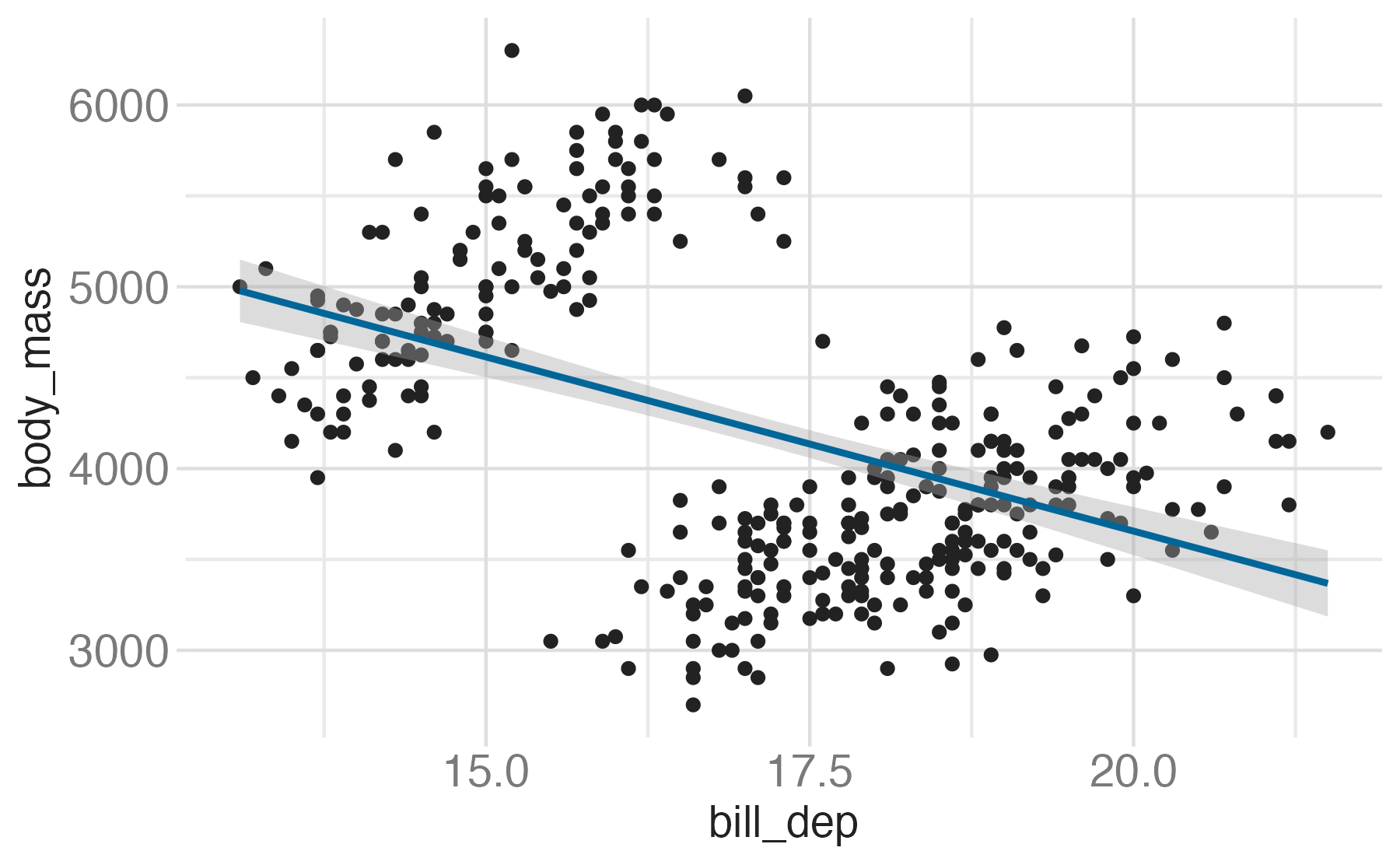
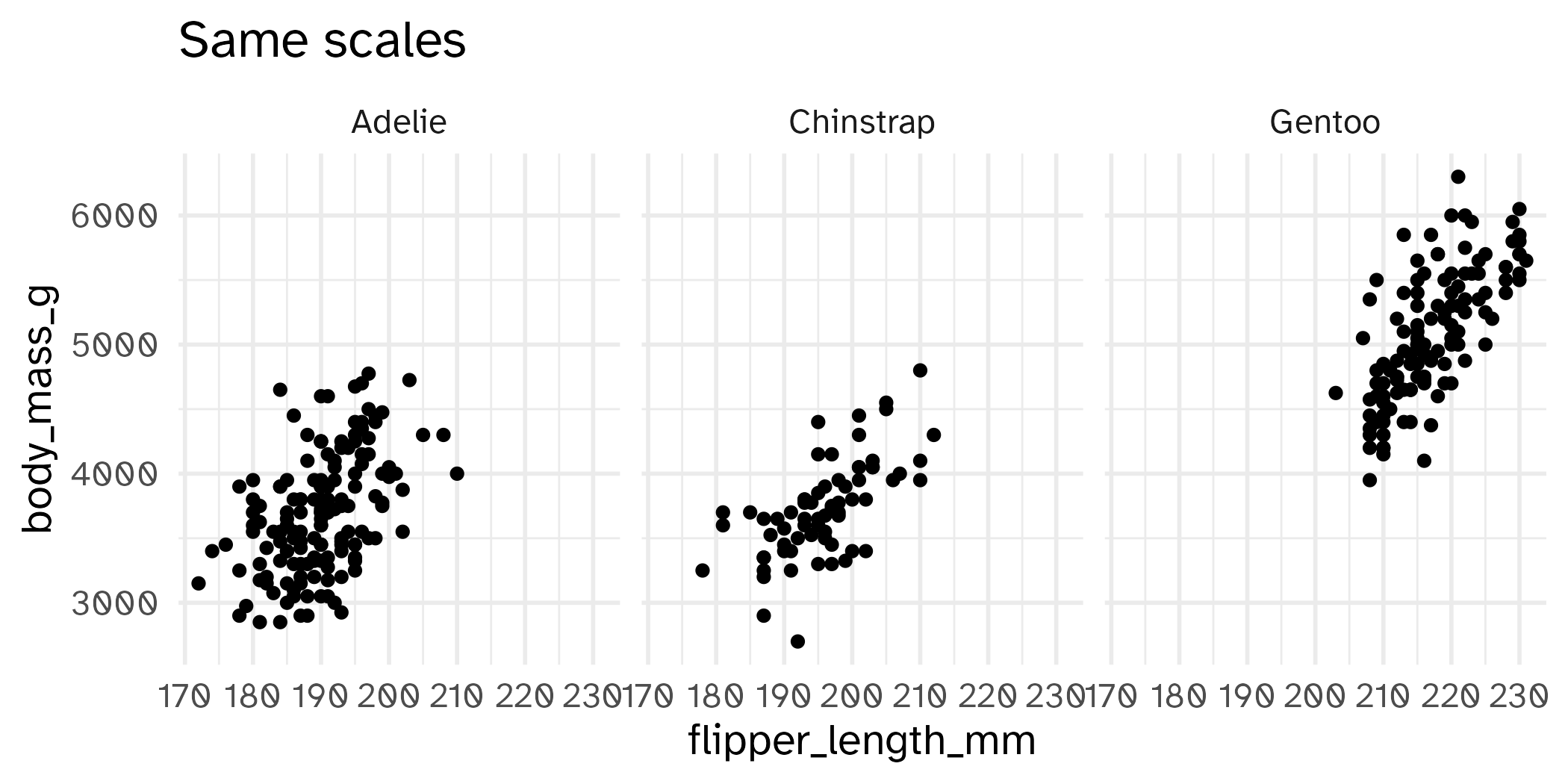
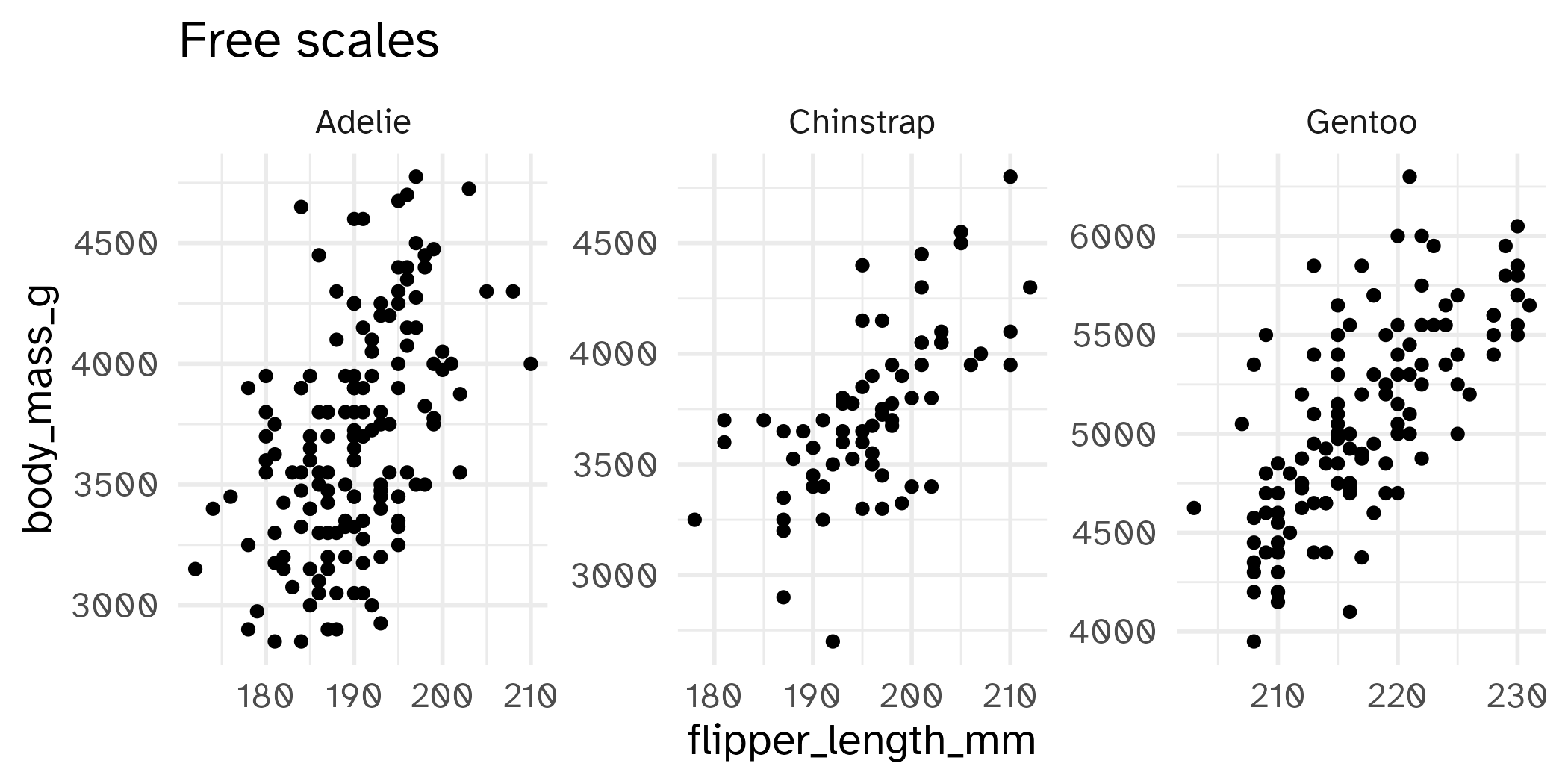
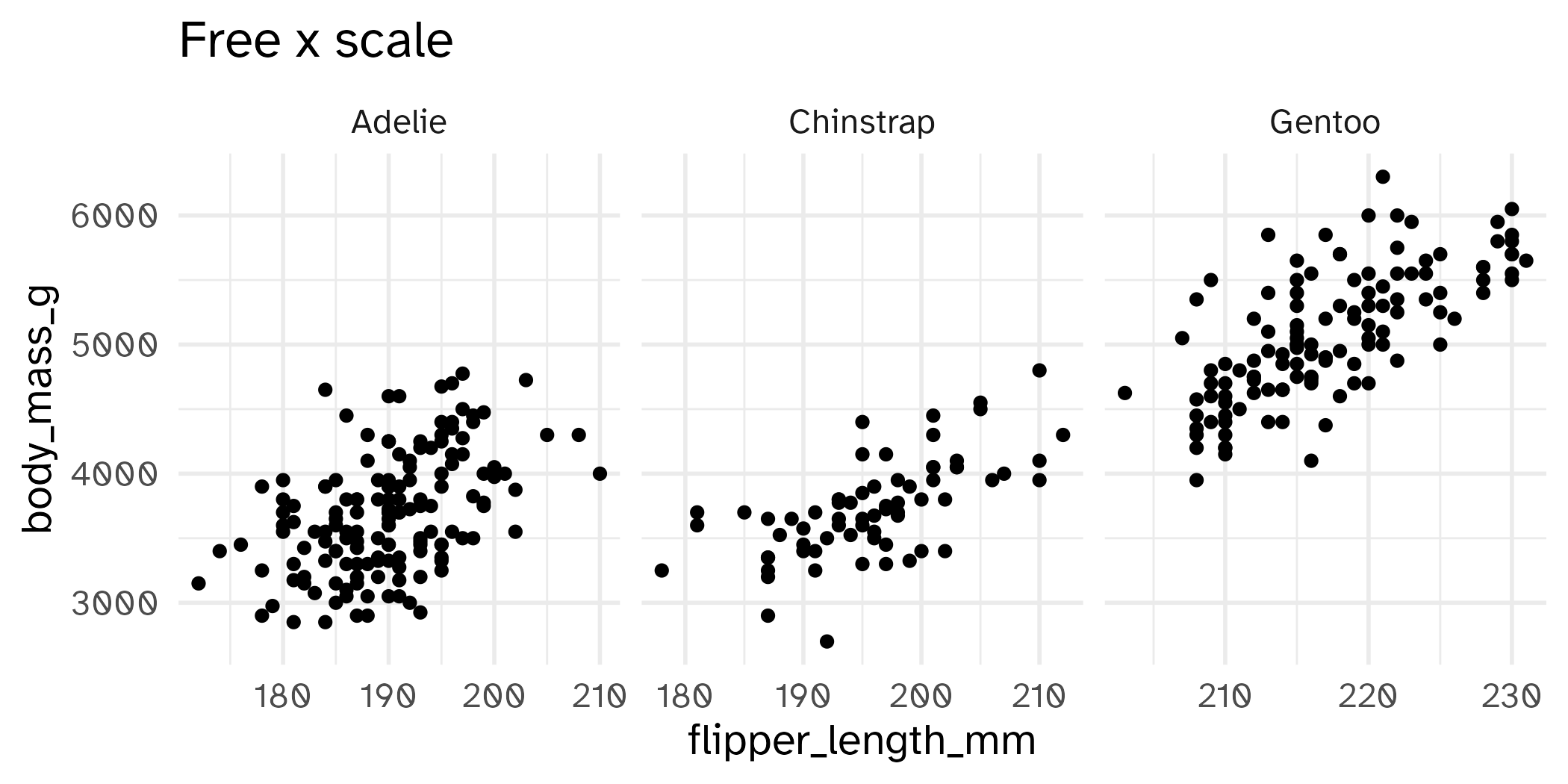
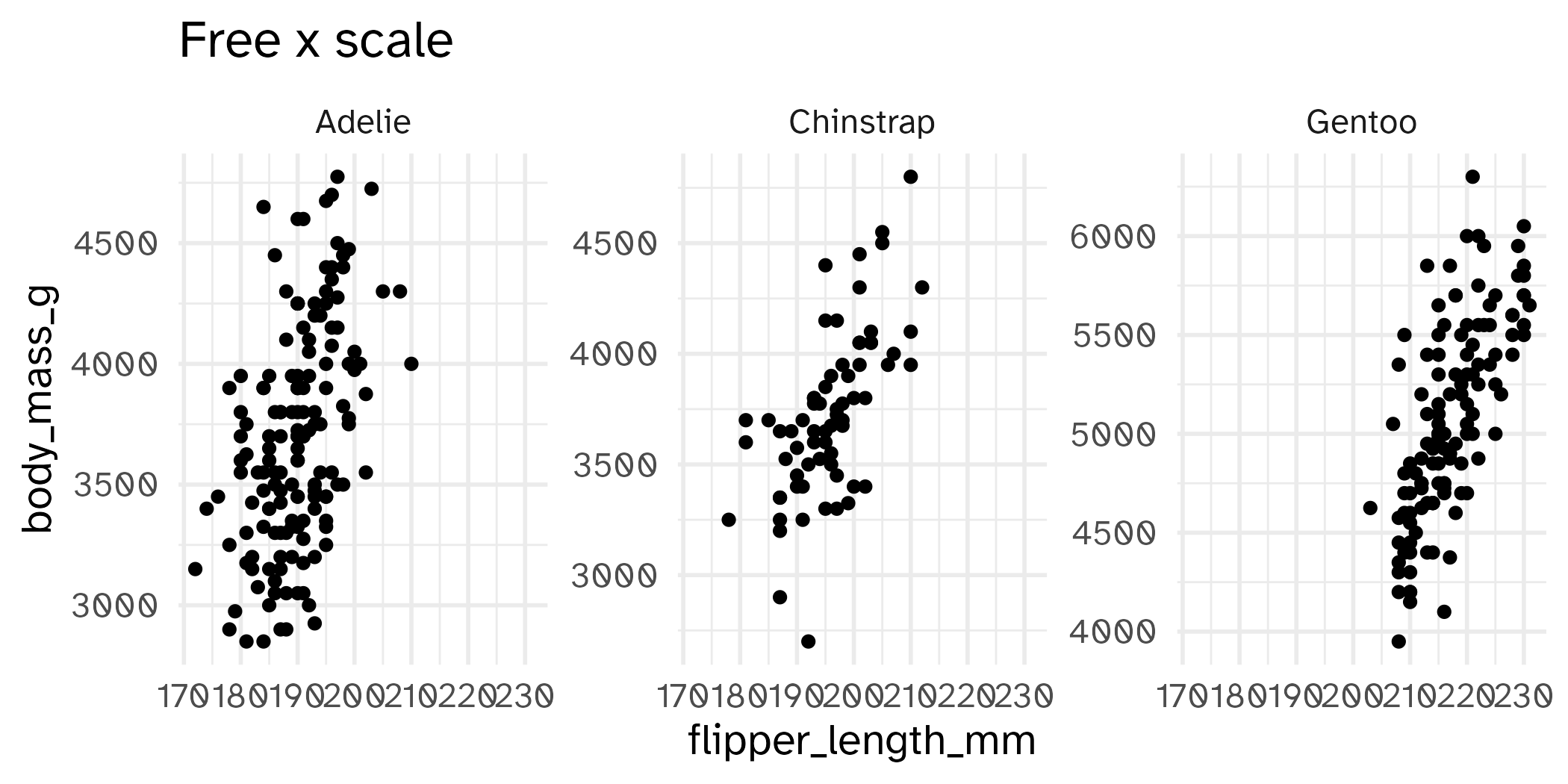
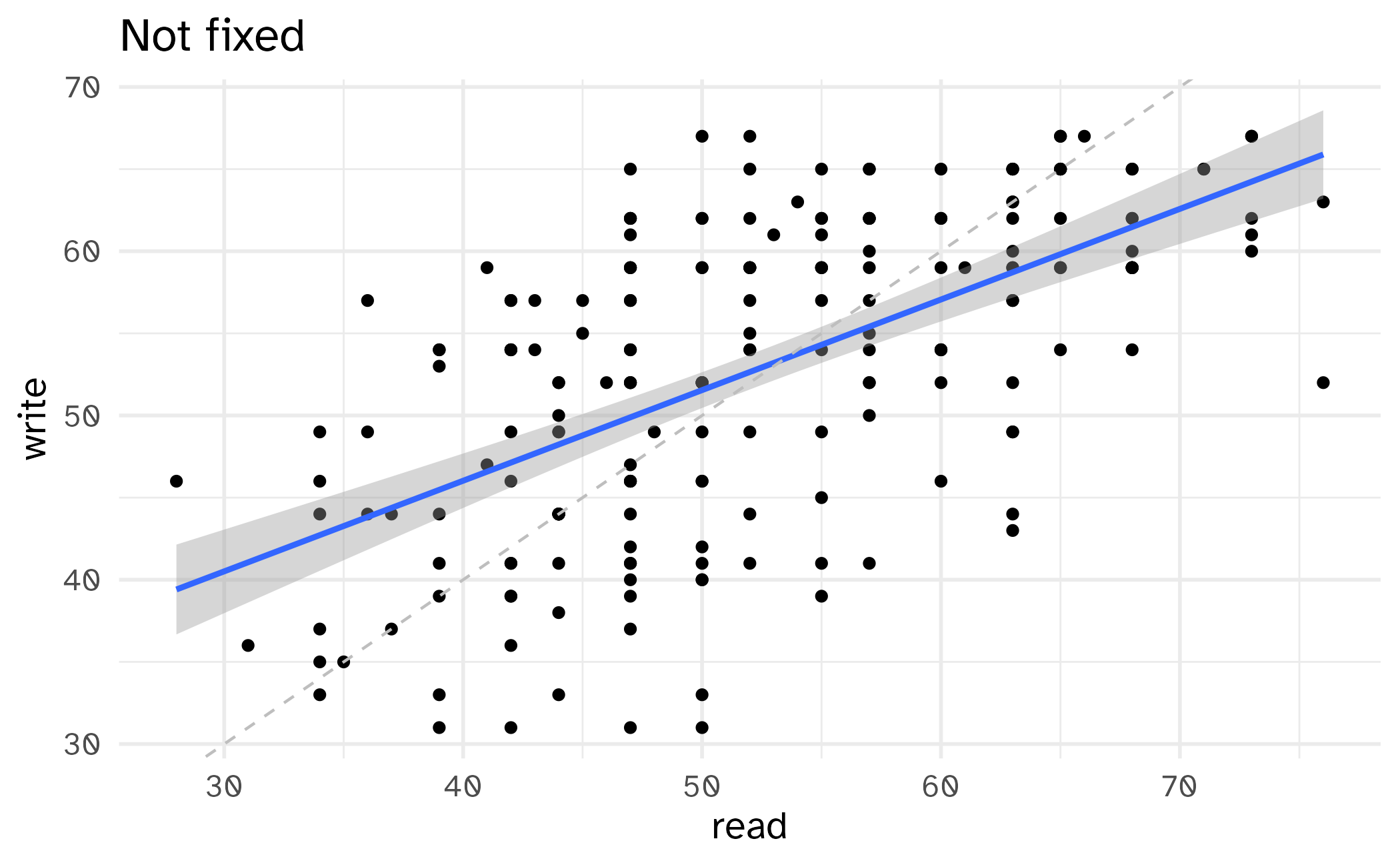
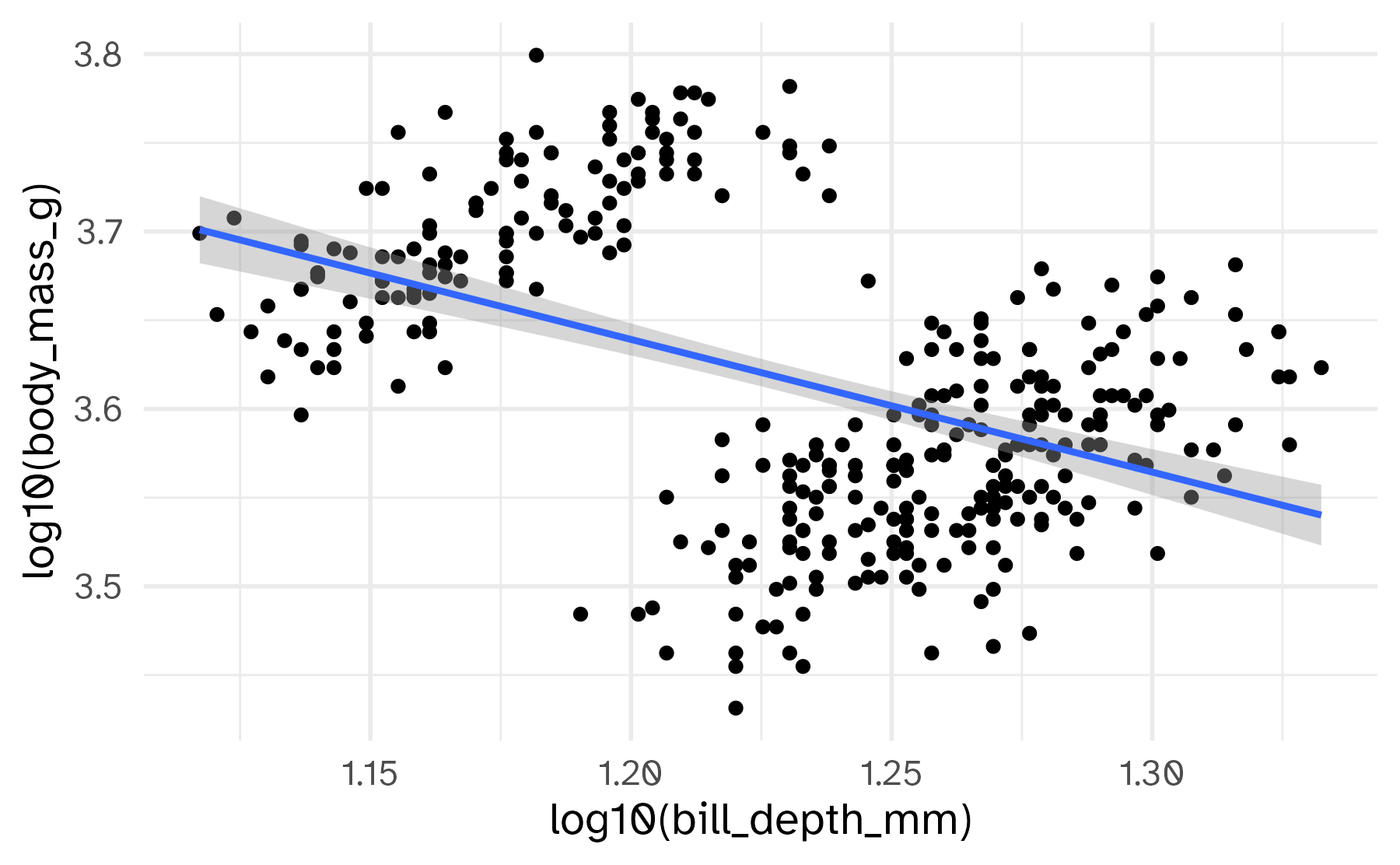
Setting limits: what the plots say
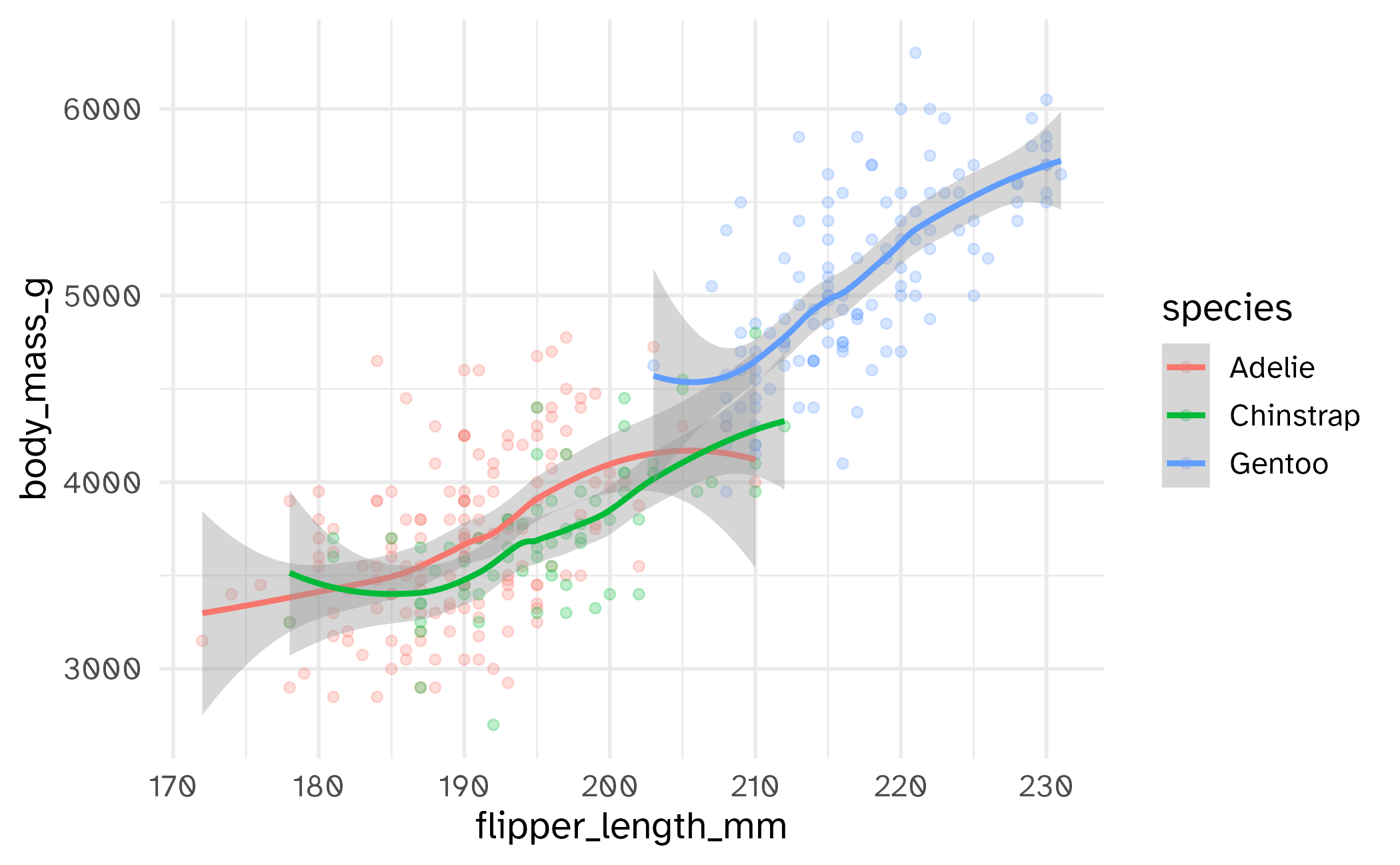
Setting limits: what the plots say
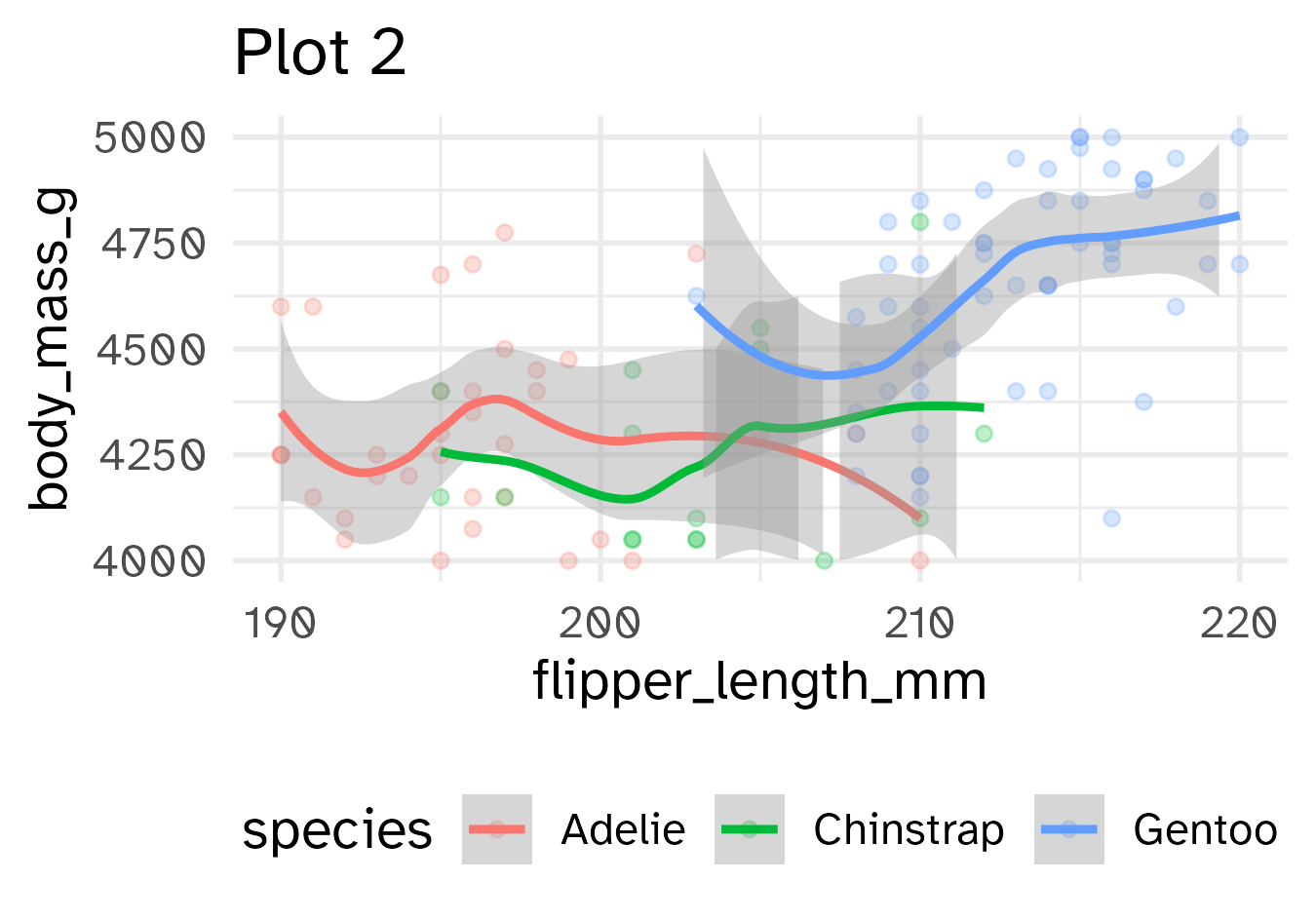
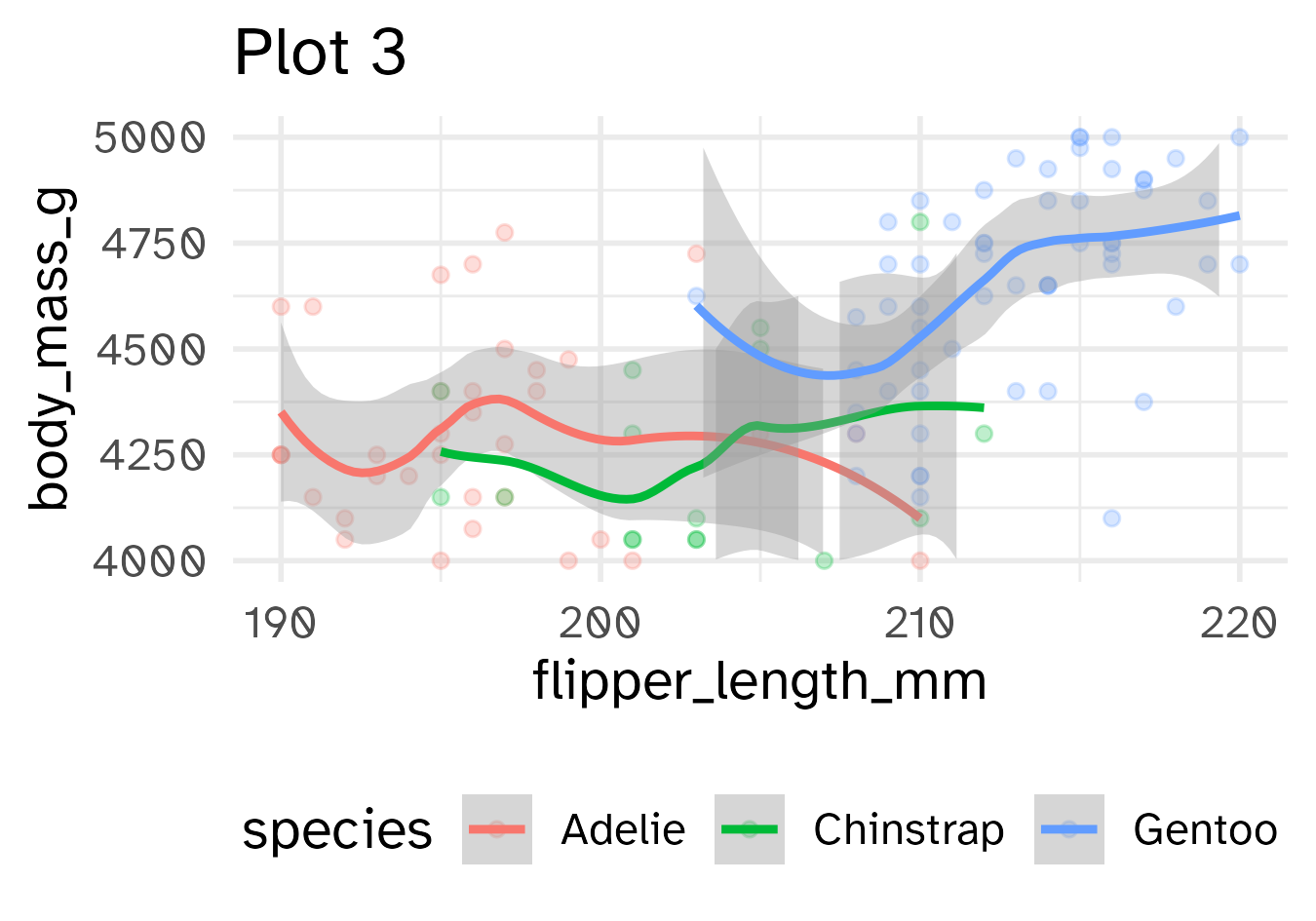
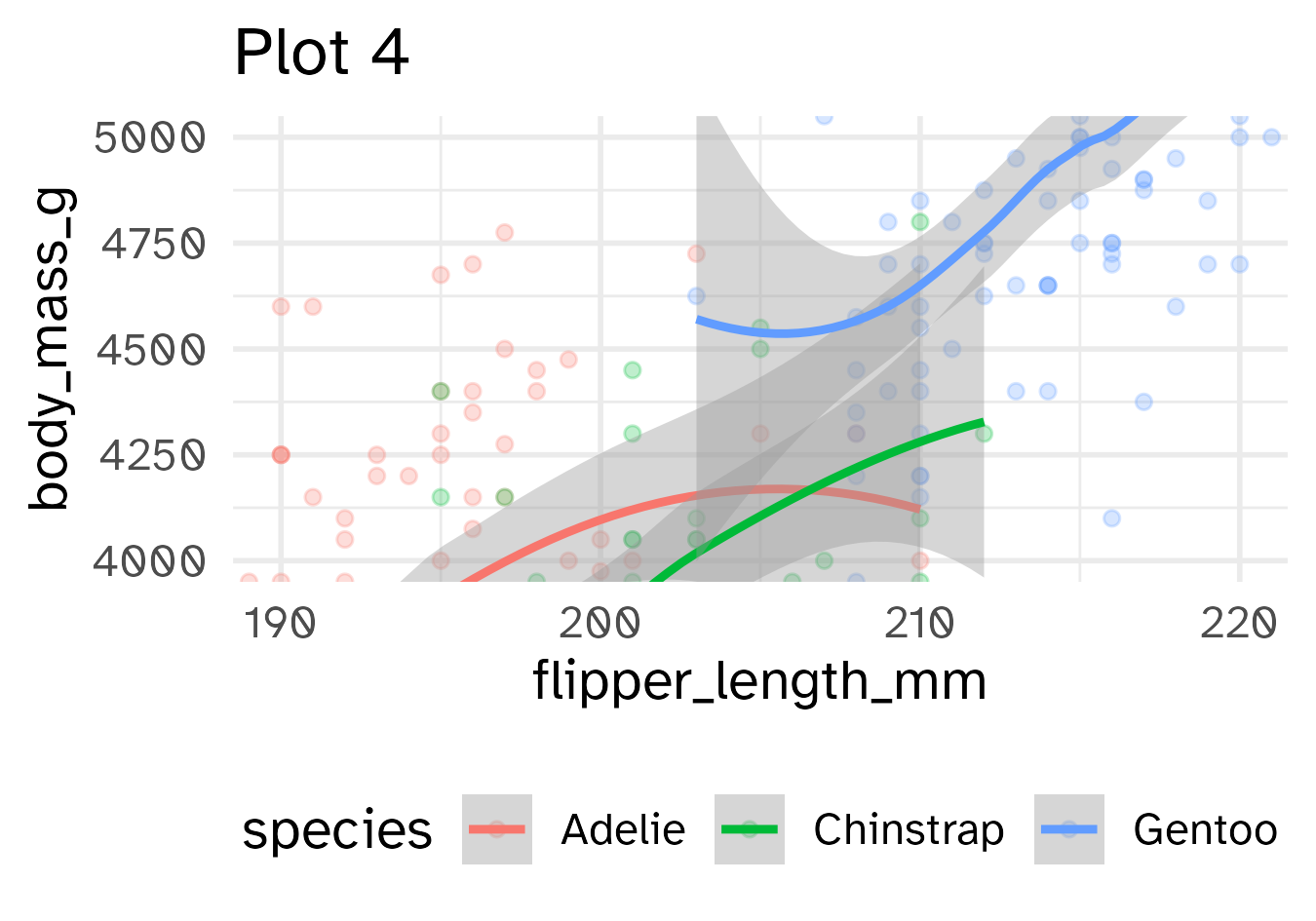
Identify the differences between each plot. Focus on the range of the x and y axes as well as the contents of the plots.
02:00
base_plot +
labs(title = "Plot 1")
base_plot +
scale_x_continuous(limits = c(190, 220)) +
scale_y_continuous(limits = c(4000, 5000)) +
labs(title = "Plot 2")
base_plot +
xlim(190, 220) +
ylim(4000, 5000) +
labs(title = "Plot 3")
base_plot +
coord_cartesian(xlim = c(190, 220),
ylim = c(4000, 5000)) +
labs(title = "Plot 4")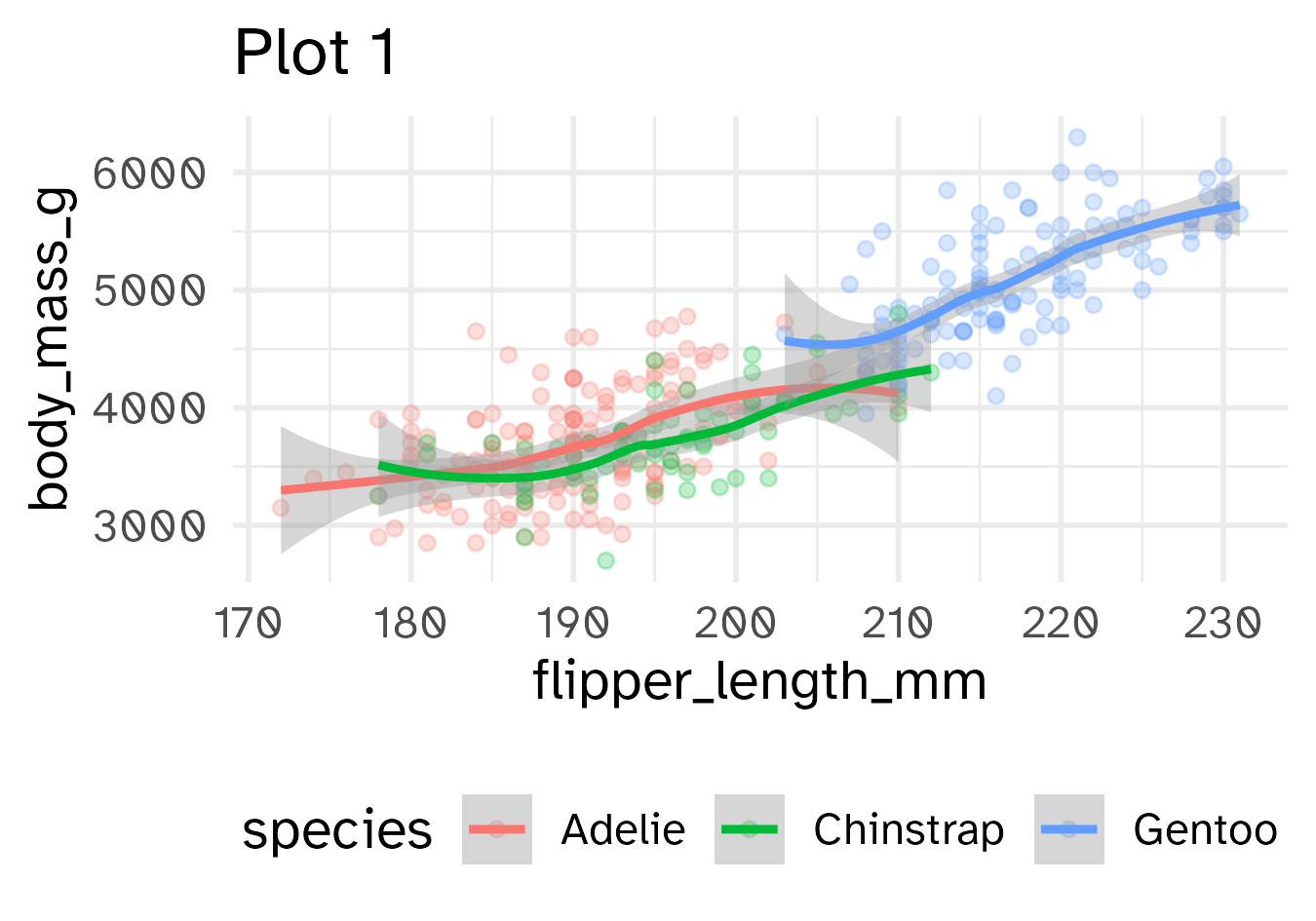
Setting limits: what the warnings say
base_plot +
labs(title = "Plot 1")
## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'
## Warning: Removed 2 rows containing non-finite outside the scale range (`stat_smooth()`).
## Warning: Removed 2 rows containing missing values or values outside the scale range
## (`geom_point()`).
base_plot +
scale_x_continuous(limits = c(190, 220)) +
scale_y_continuous(limits = c(4000, 5000)) +
labs(title = "Plot 2")
## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'
## Warning: Removed 235 rows containing non-finite outside the scale range (`stat_smooth()`).
## Warning: Removed 235 rows containing missing values or values outside the scale range
## (`geom_point()`).
base_plot +
xlim(190, 220) +
ylim(4000, 5000) +
labs(title = "Plot 3")
## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'
## Warning: Removed 235 rows containing non-finite outside the scale range (`stat_smooth()`).
## Removed 235 rows containing missing values or values outside the scale range (`geom_point()`).
base_plot +
coord_cartesian(xlim = c(190, 220),
ylim = c(4000, 5000)) +
labs(title = "Plot 4")
## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'
## Warning: Removed 2 rows containing non-finite outside the scale range (`stat_smooth()`).
## Warning: Removed 2 rows containing missing values or values outside the scale range
## (`geom_point()`).Setting limits
- Setting scale limits: Any data outside the limits is thrown away
scale_*_continuous(limits = ...)xlim()andylim()
- Setting coordinate system limits: Use all the data, but only display a small region of the plot (zooming in)
coord_cartesian(xlim = ..., ylim = ...)
Cropping scatterplot
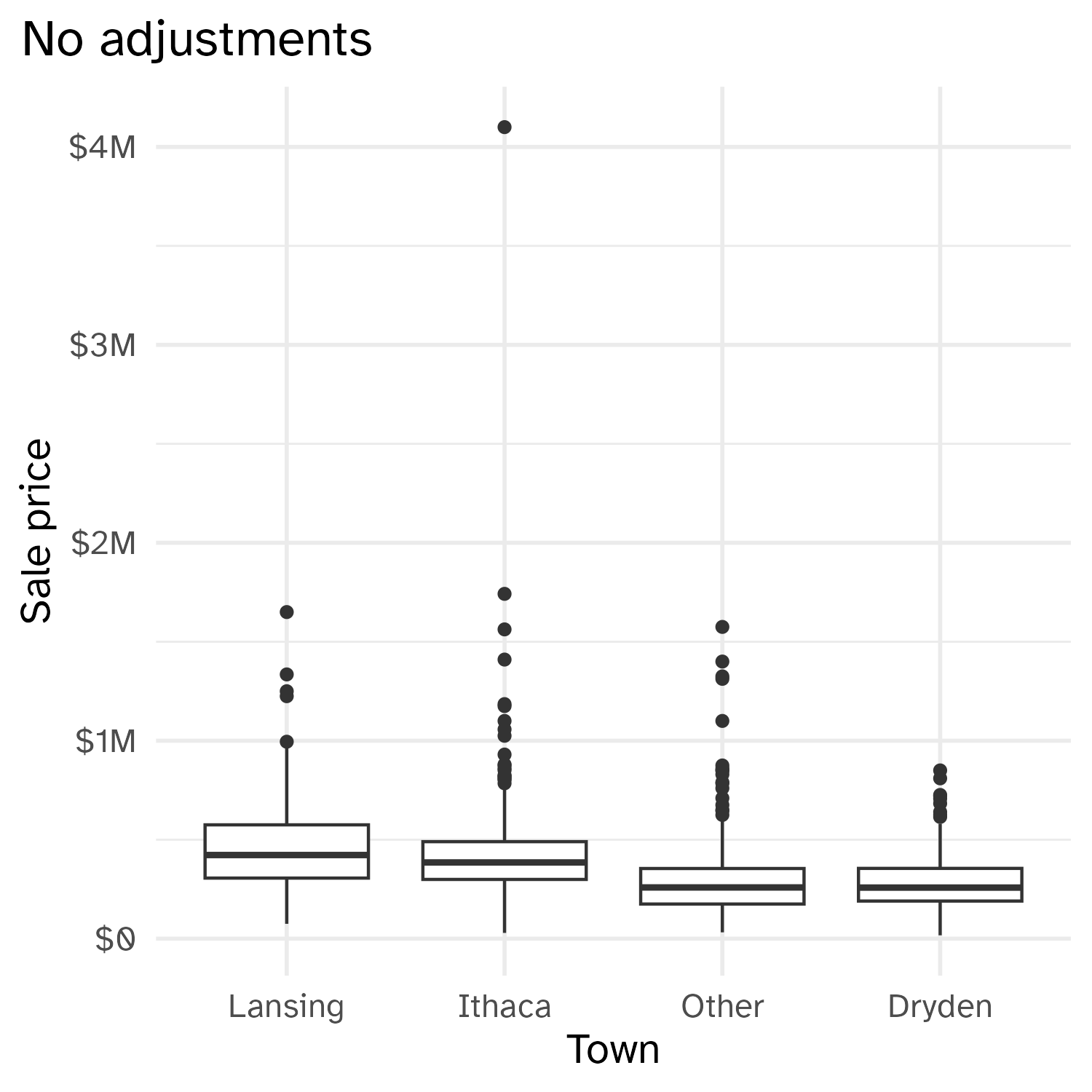
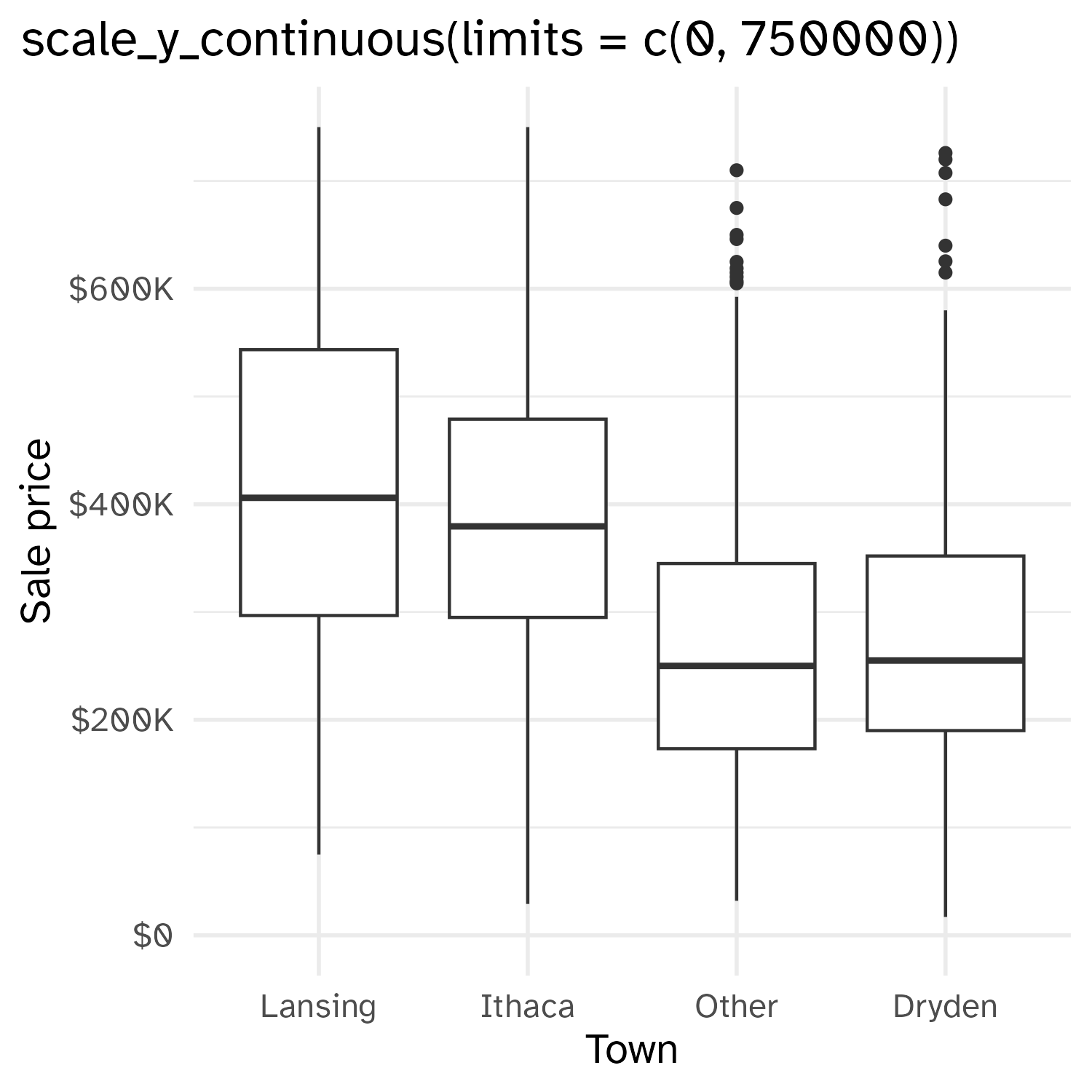
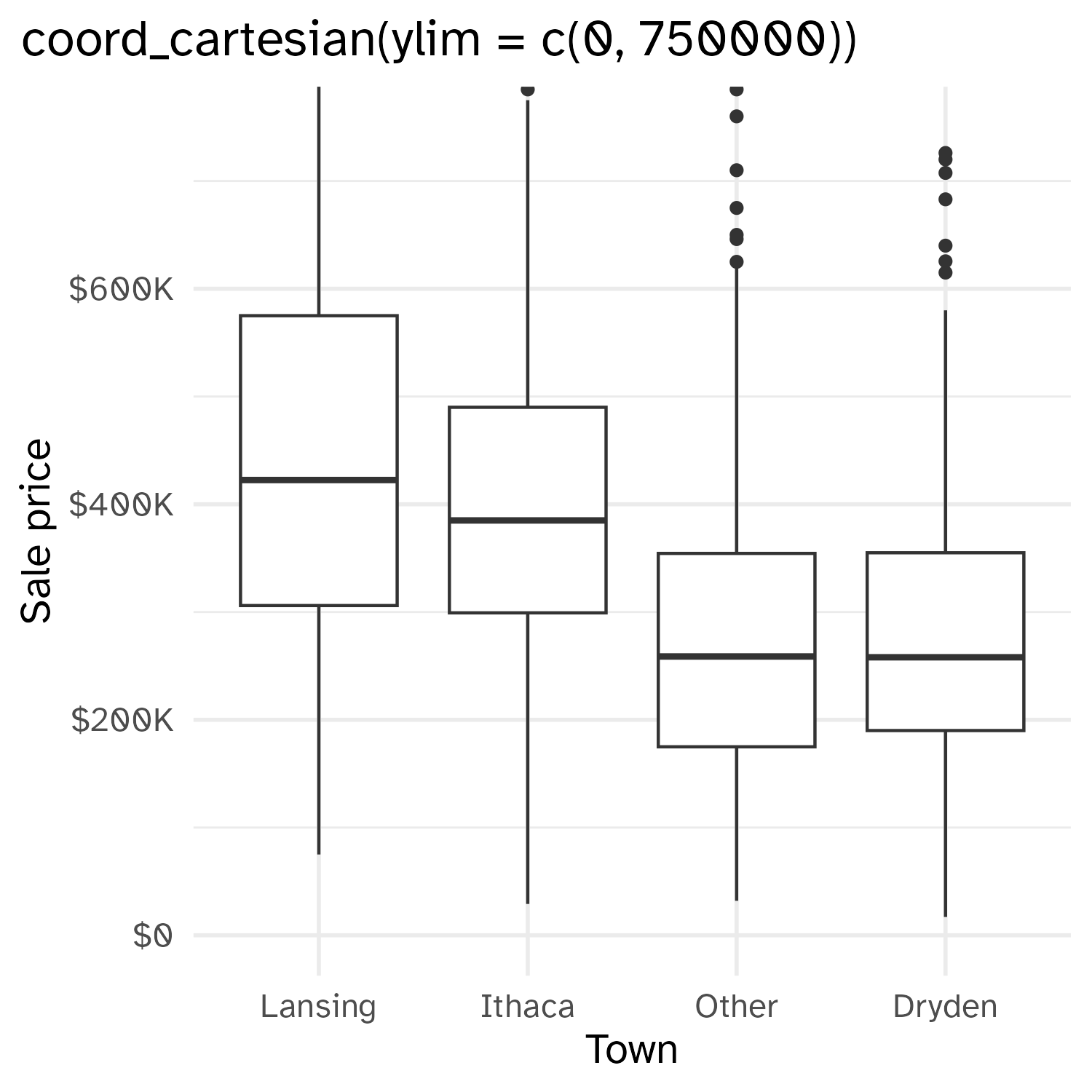
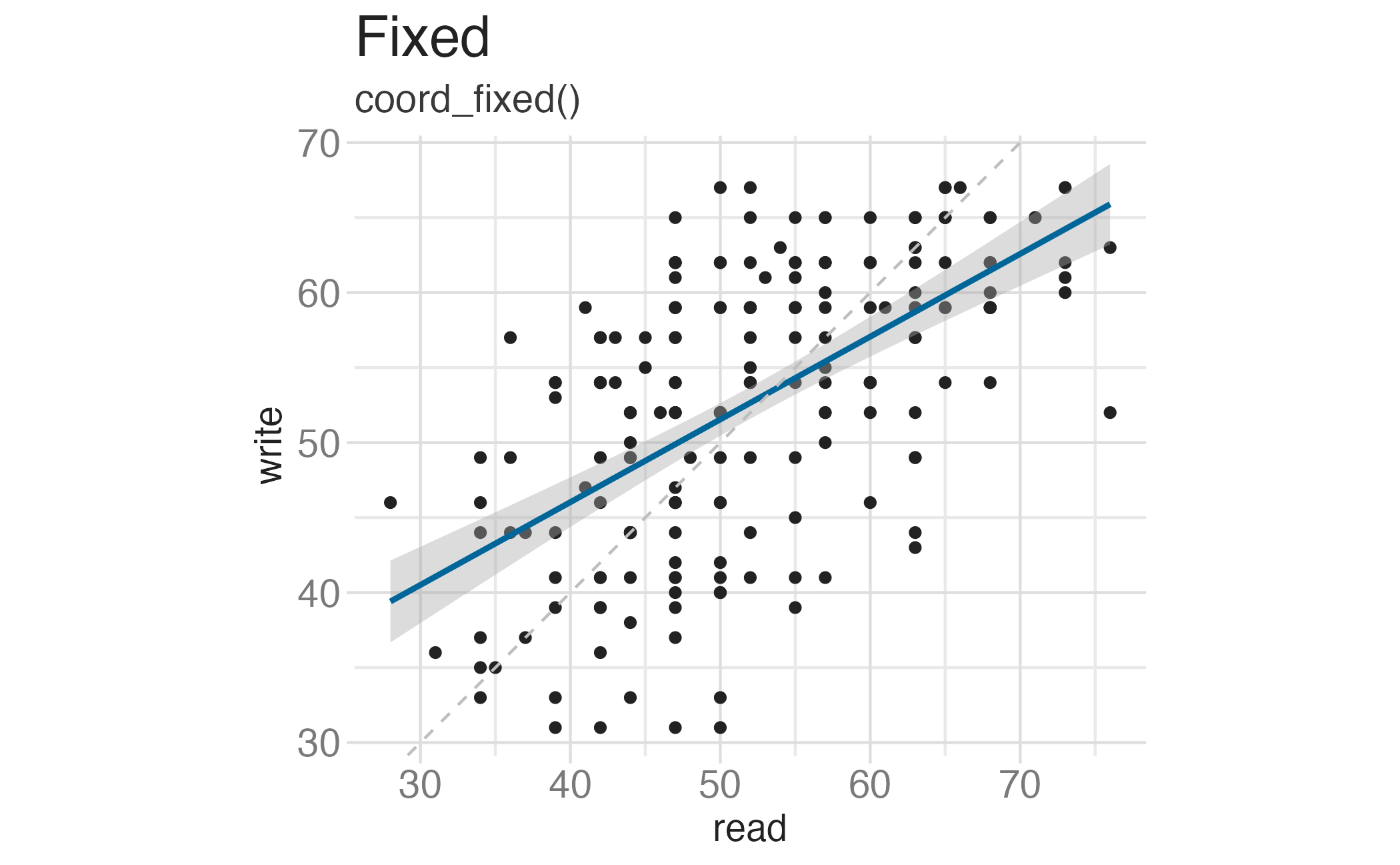
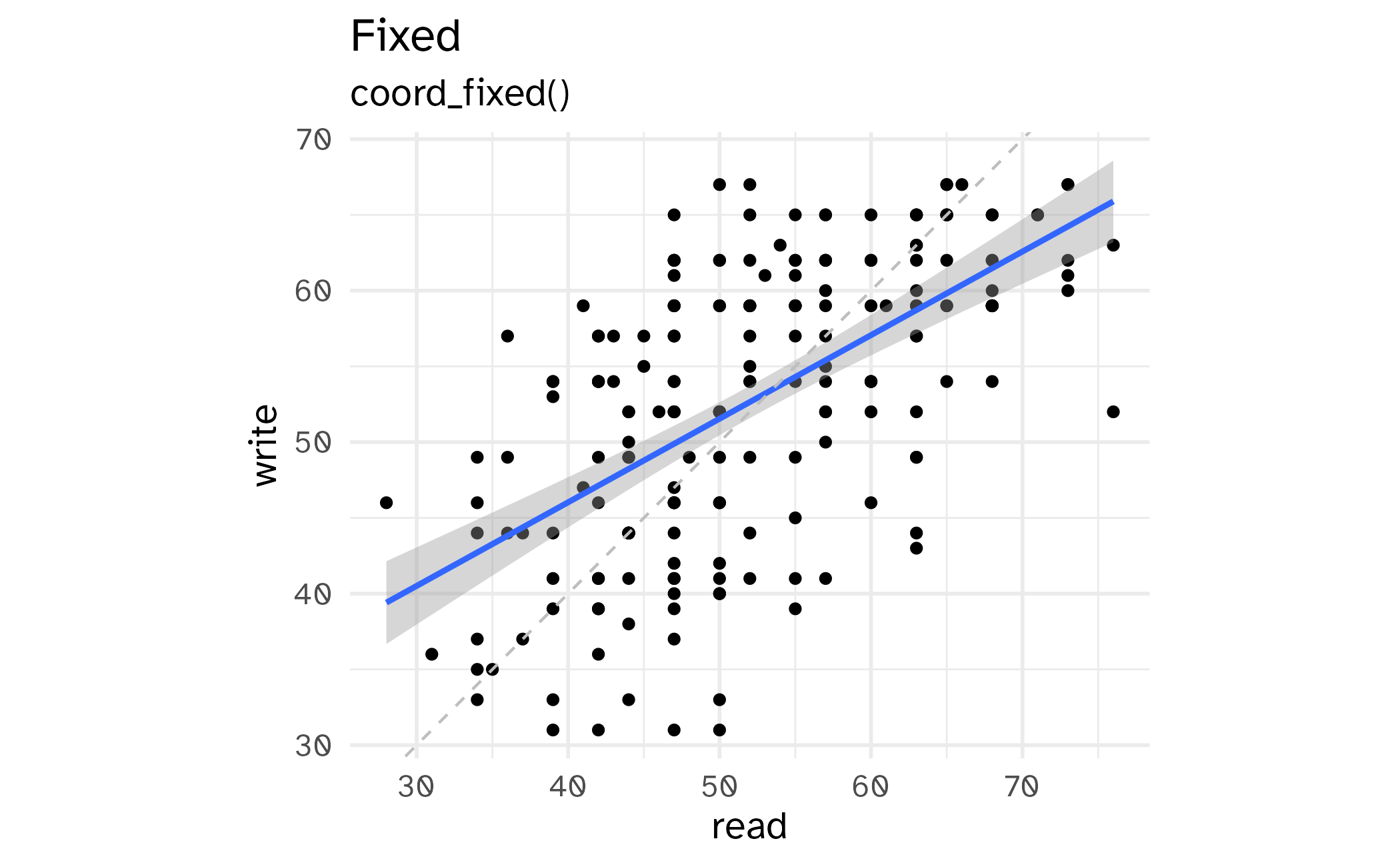
Fixing aspect ratio with coord_cartesian(ratio = X)
Useful when having a fixed aspect ratio makes sense, e.g. scores on two tests (reading and writing) on the same scale (0 to 100 points)
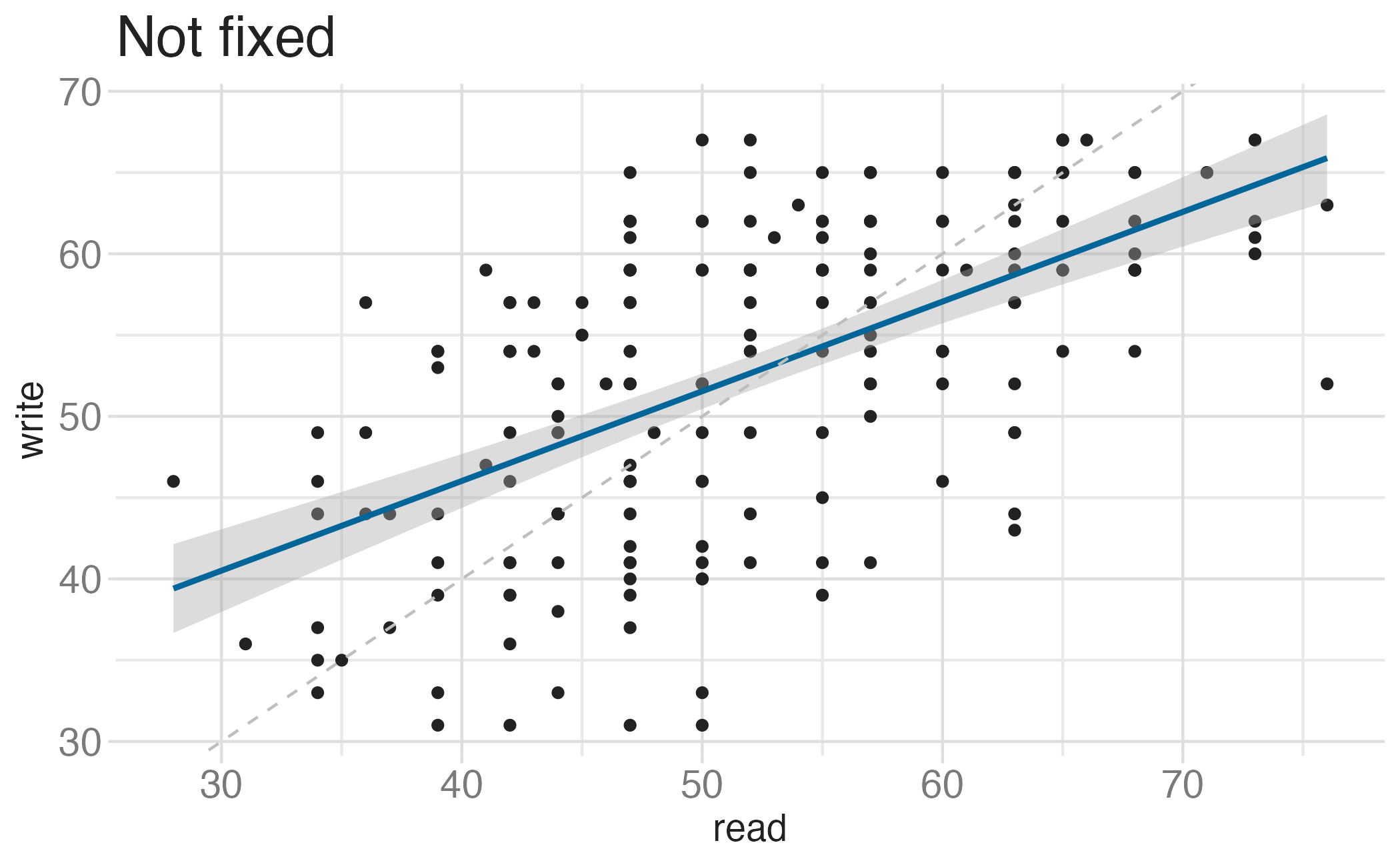
Transformations
Polar coordinate systems
Radial charts with coord_radial()
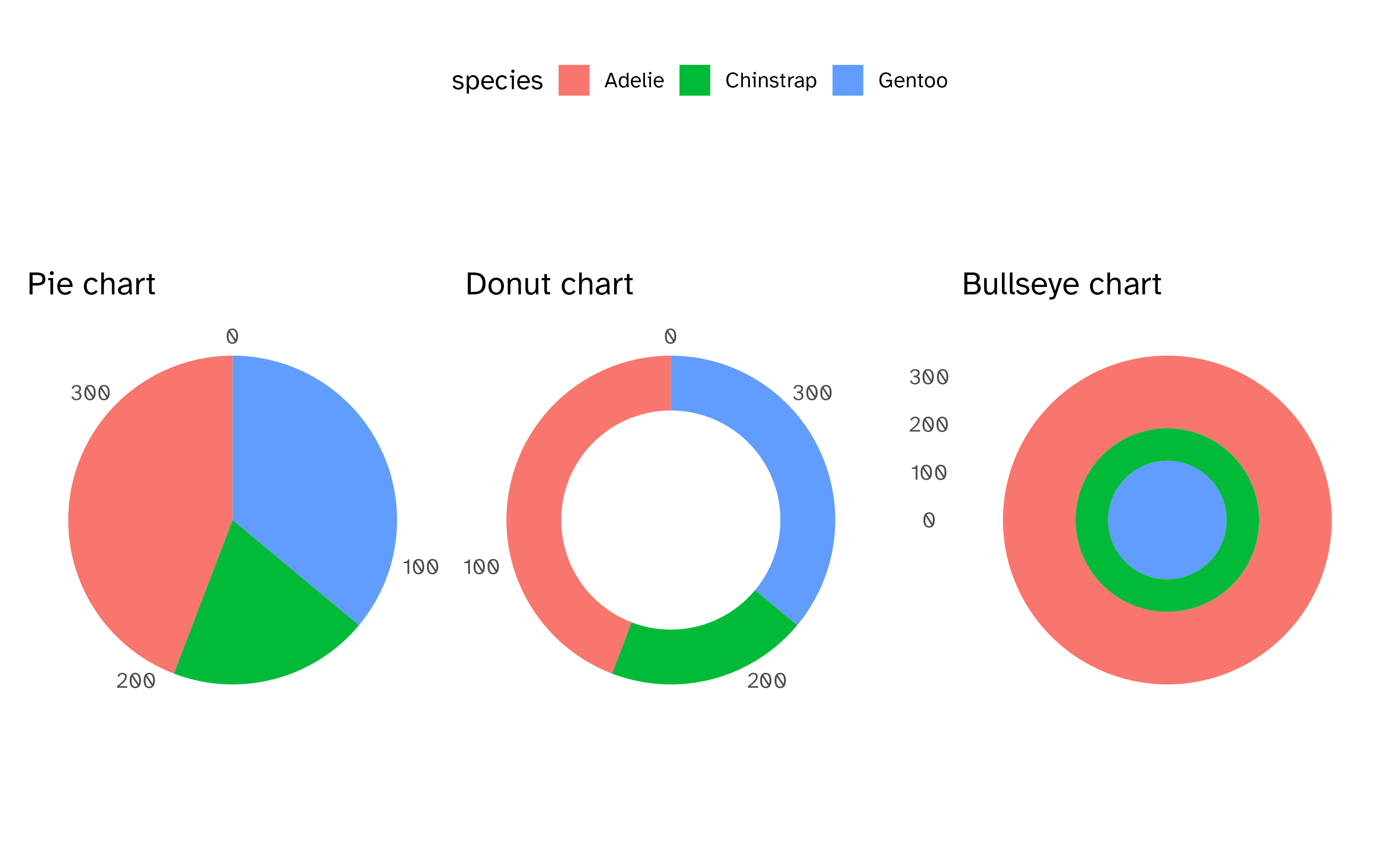
Circular bar charts
More examples: R Graph Gallery
Circular lollipop charts
Source: R Graph Gallery
Authentic pie chart
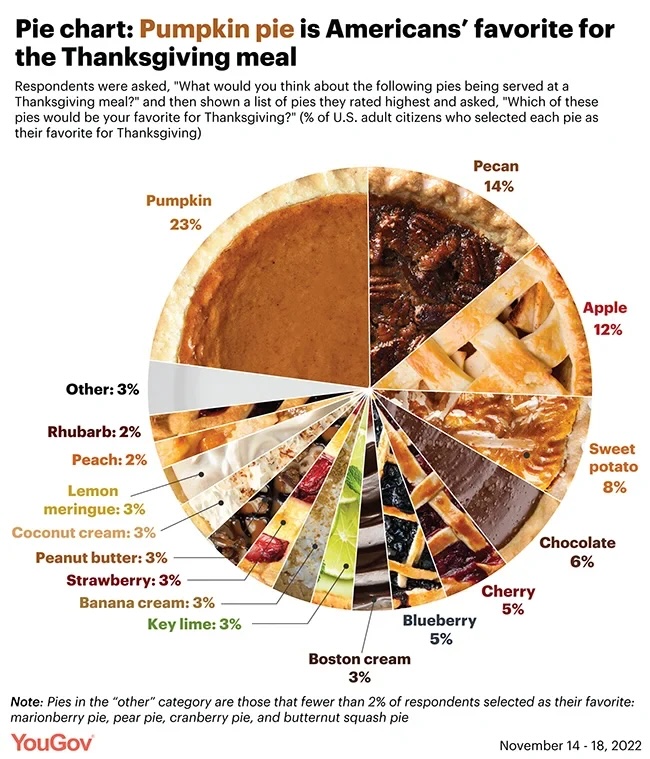
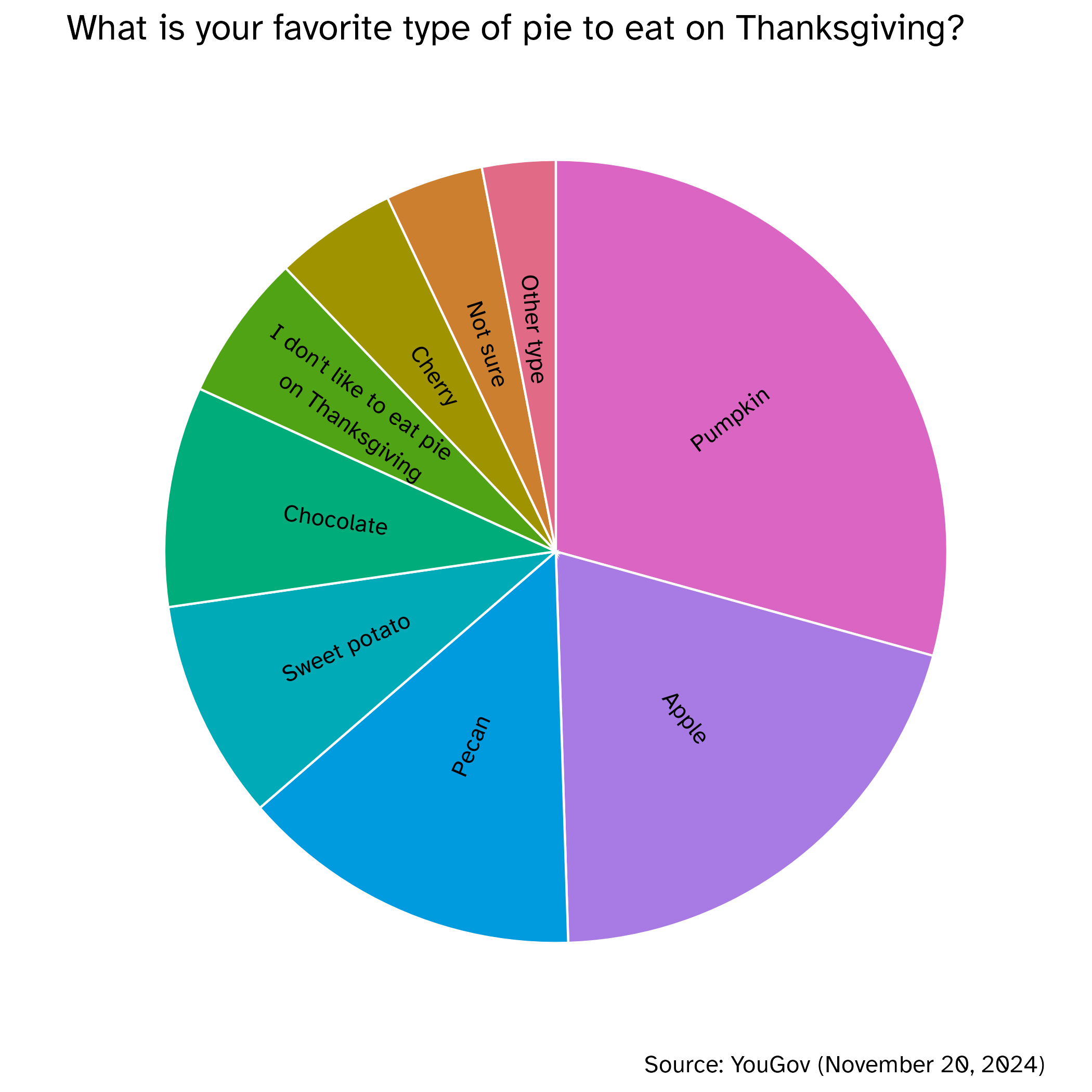
Image credit: YouGov
Pie charts
What do you know about pie charts and data visualization best practices? Love ’em or lose ’em?
Pie charts
For categorical variables with few levels, pie charts can work well
For categorical variables with many levels, pie charts are difficult to read
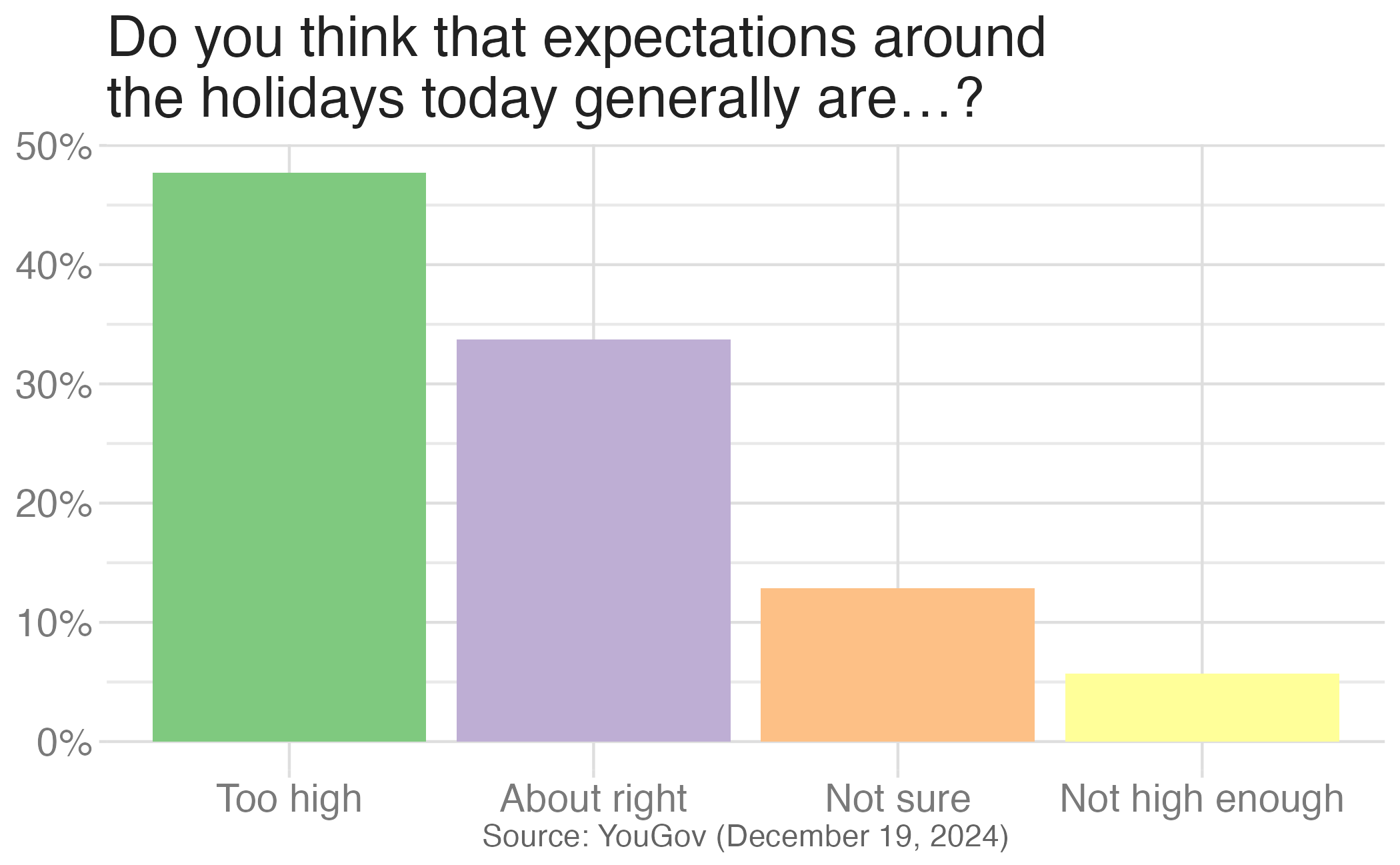
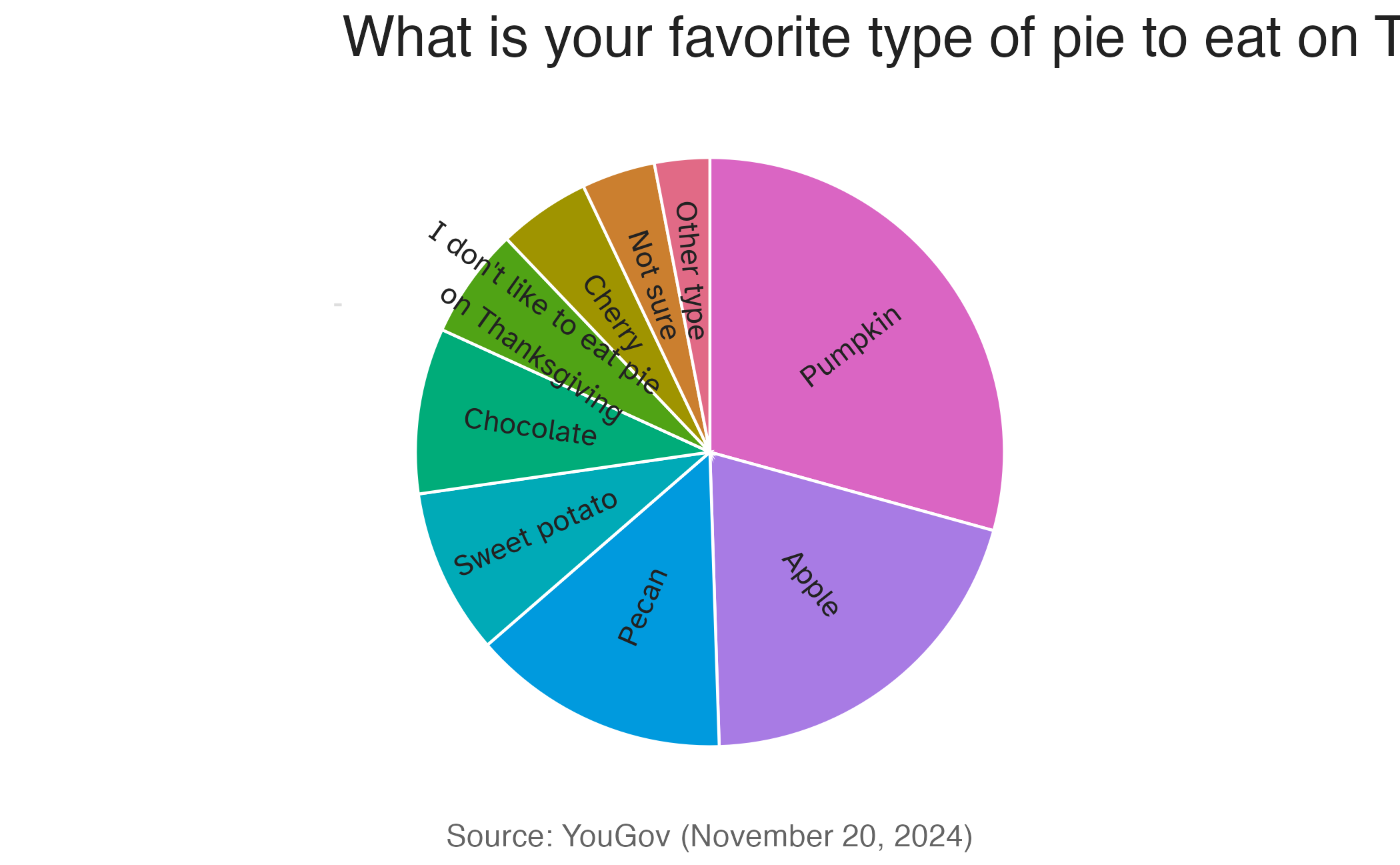
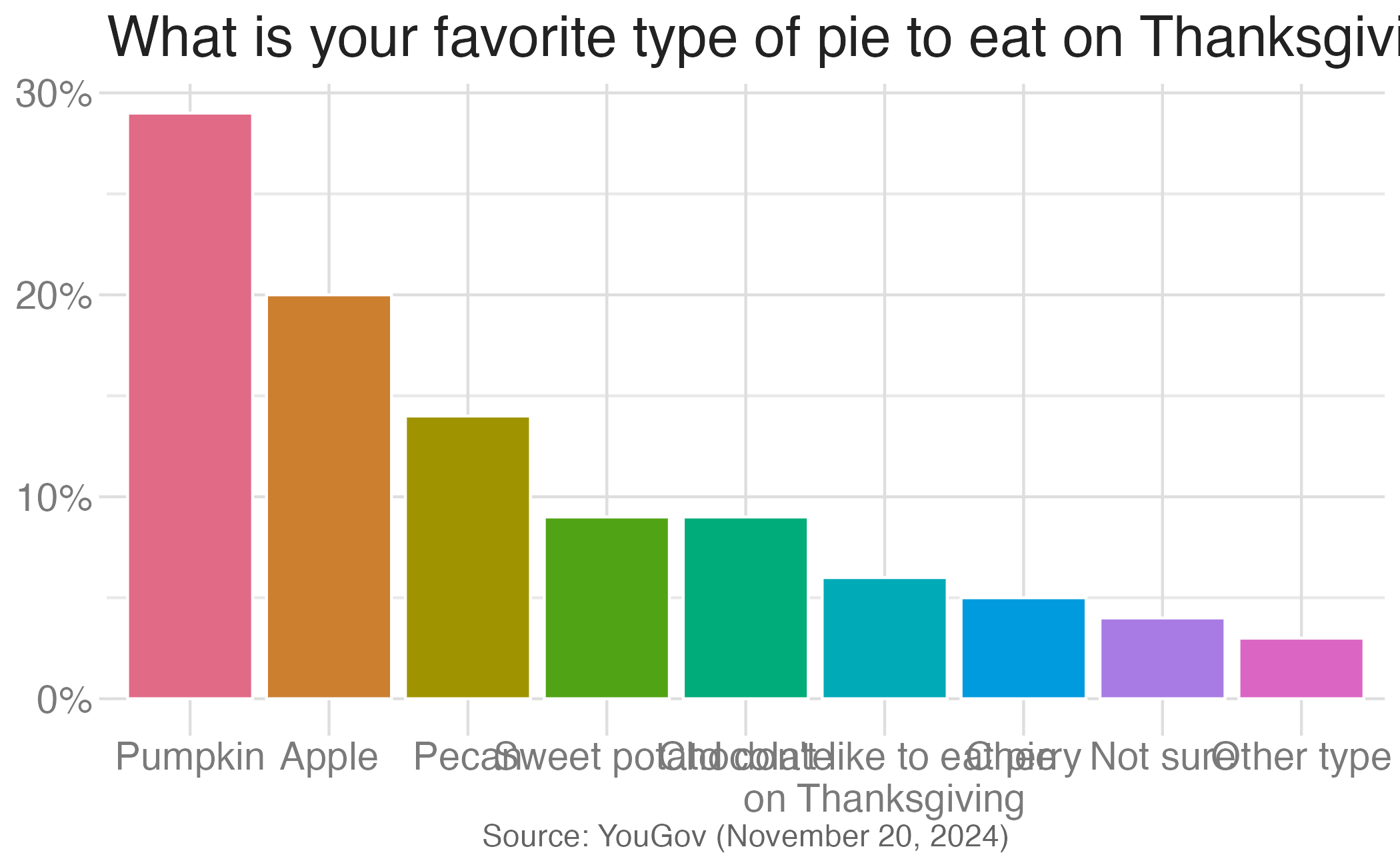

Waffle charts
- Like with pie charts, work best when the number of levels represented is low
- Unlike pie charts, easier to compare proportions that represent non-simple fractions
Application exercise
ae-05
Instructions
- Go to the course GitHub org and find your
ae-05(repo name will be suffixed with your GitHub name). - Clone the repo in Positron, run
renv::restore()to install the required packages, open the Quarto document in the repo, and follow along and complete the exercises. - Render, commit, and push your edits by the AE deadline – end of the day
12:00
Facets
facet_*()
facet_wrap()- “wraps” a 1d ribbon of panels into 2d
- generally for faceting by a single variable
facet_grid()for faceting- produces a 2d grid of panels defined by variables which form the rows and columns
- generally for faceting by two variables
facet_null(): a single plot, the default

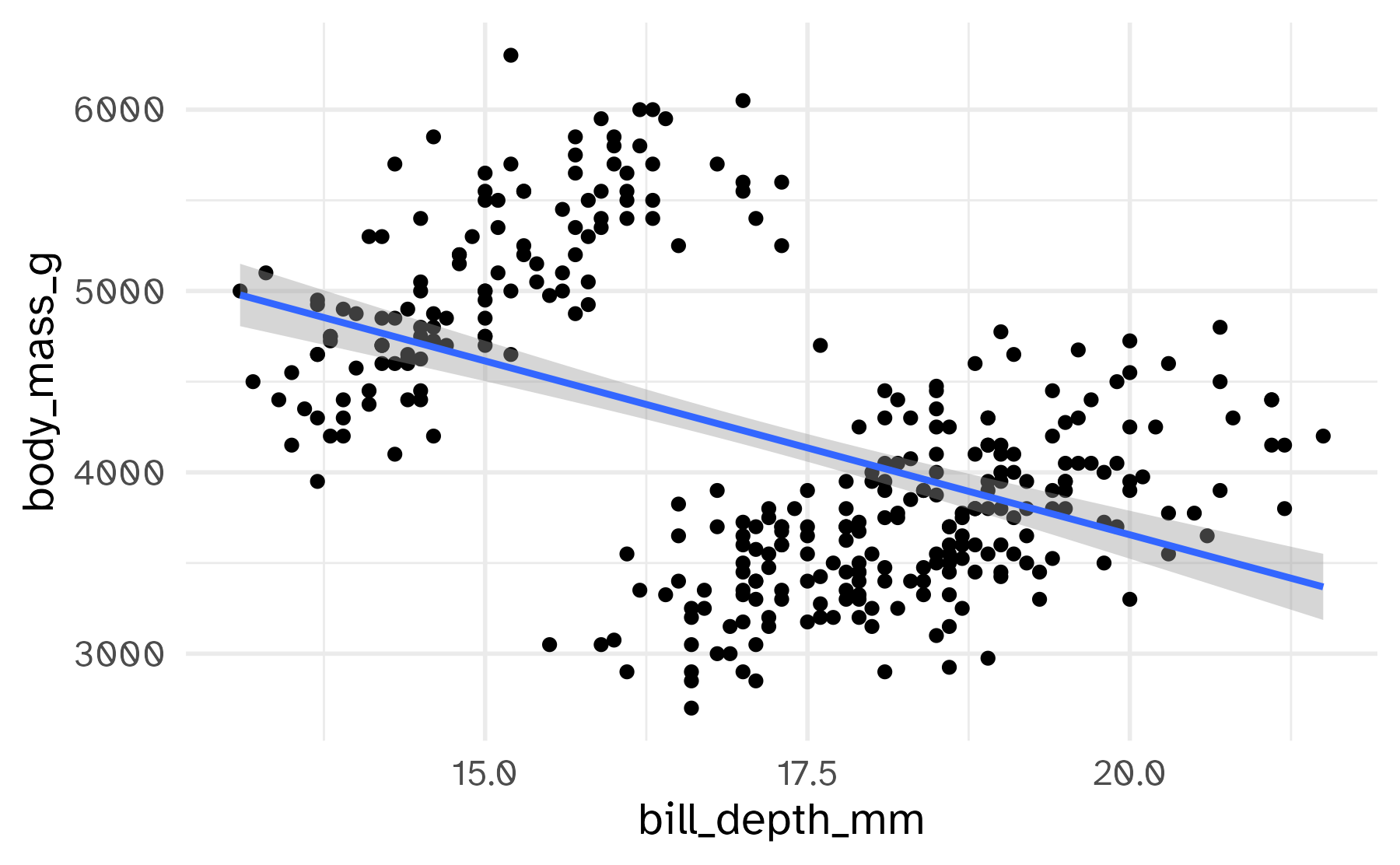
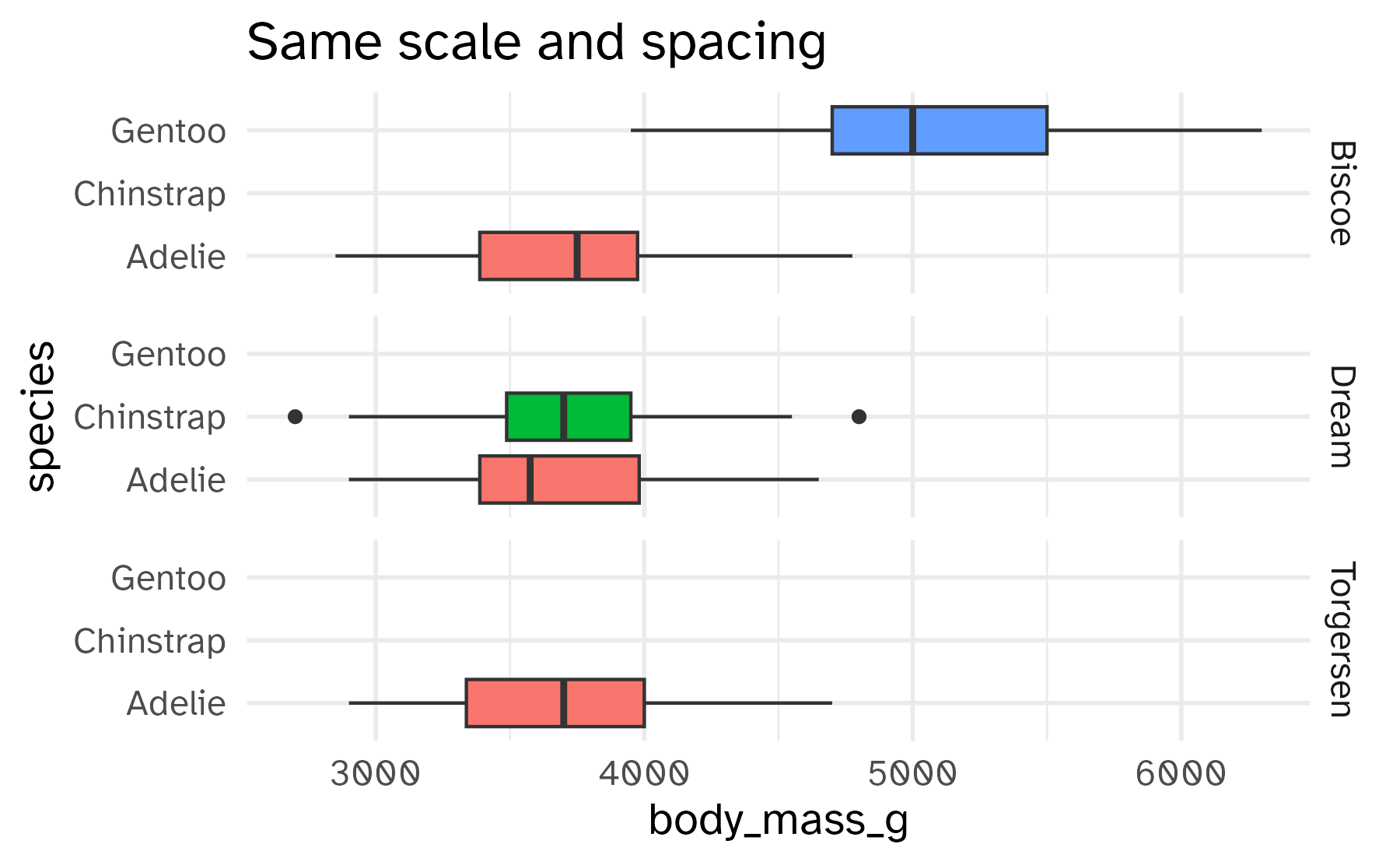
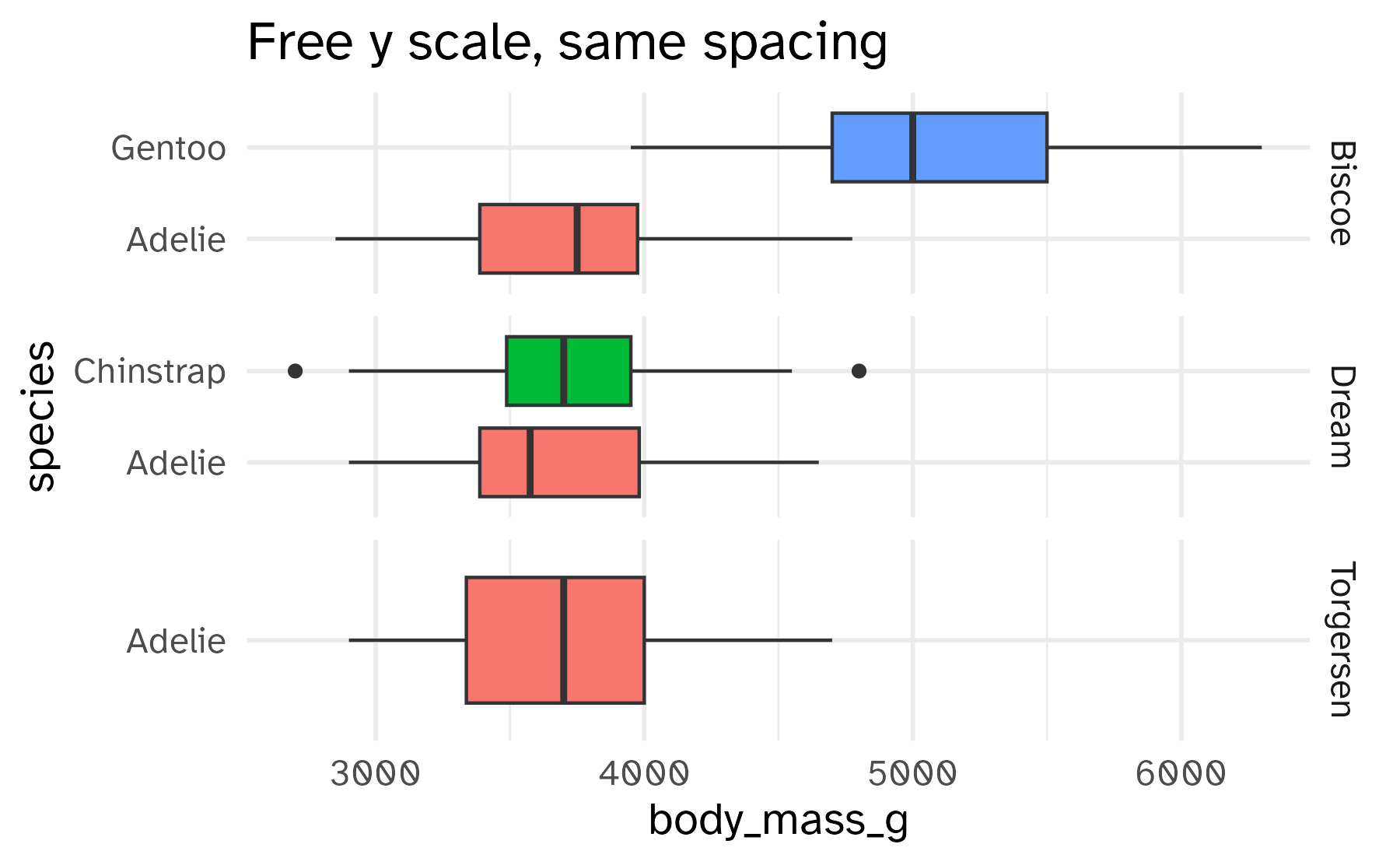
Free the scales!
Free some scales
Freeing the y scale improves the display, but it’s still not satisfying. What’s wrong with it?
ggplot(penguins, aes(y = species, x = body_mass, fill = species)) +
geom_boxplot(show.legend = FALSE) +
facet_grid(rows = vars(island)) +
labs(title = "Same scale and spacing")
ggplot(penguins, aes(y = species, x = body_mass, fill = species)) +
geom_boxplot(show.legend = FALSE) +
facet_grid(rows = vars(island), scales = "free_y") +
labs(title = "Free y scale, same spacing")Free spaces
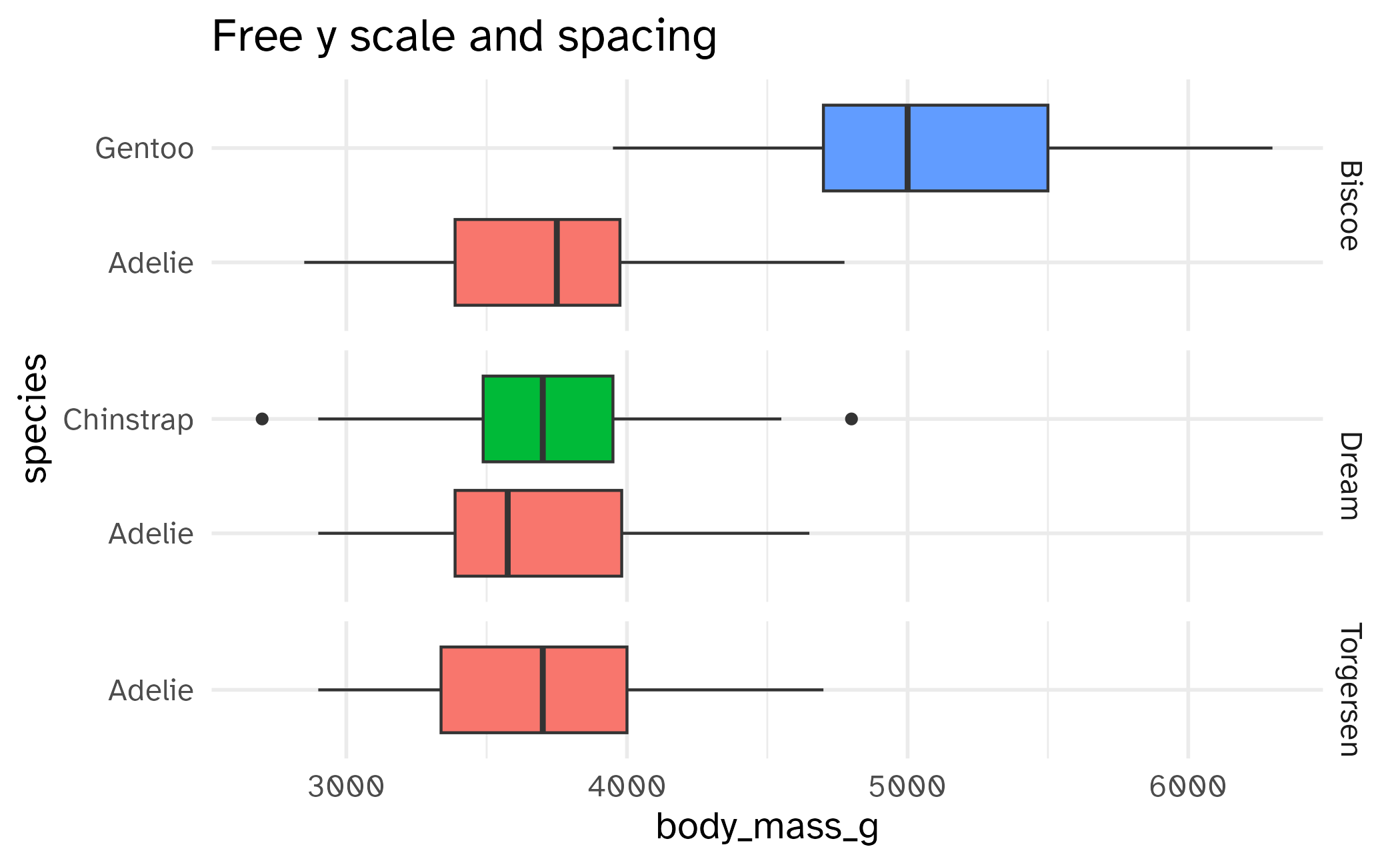
Highlighting across facets
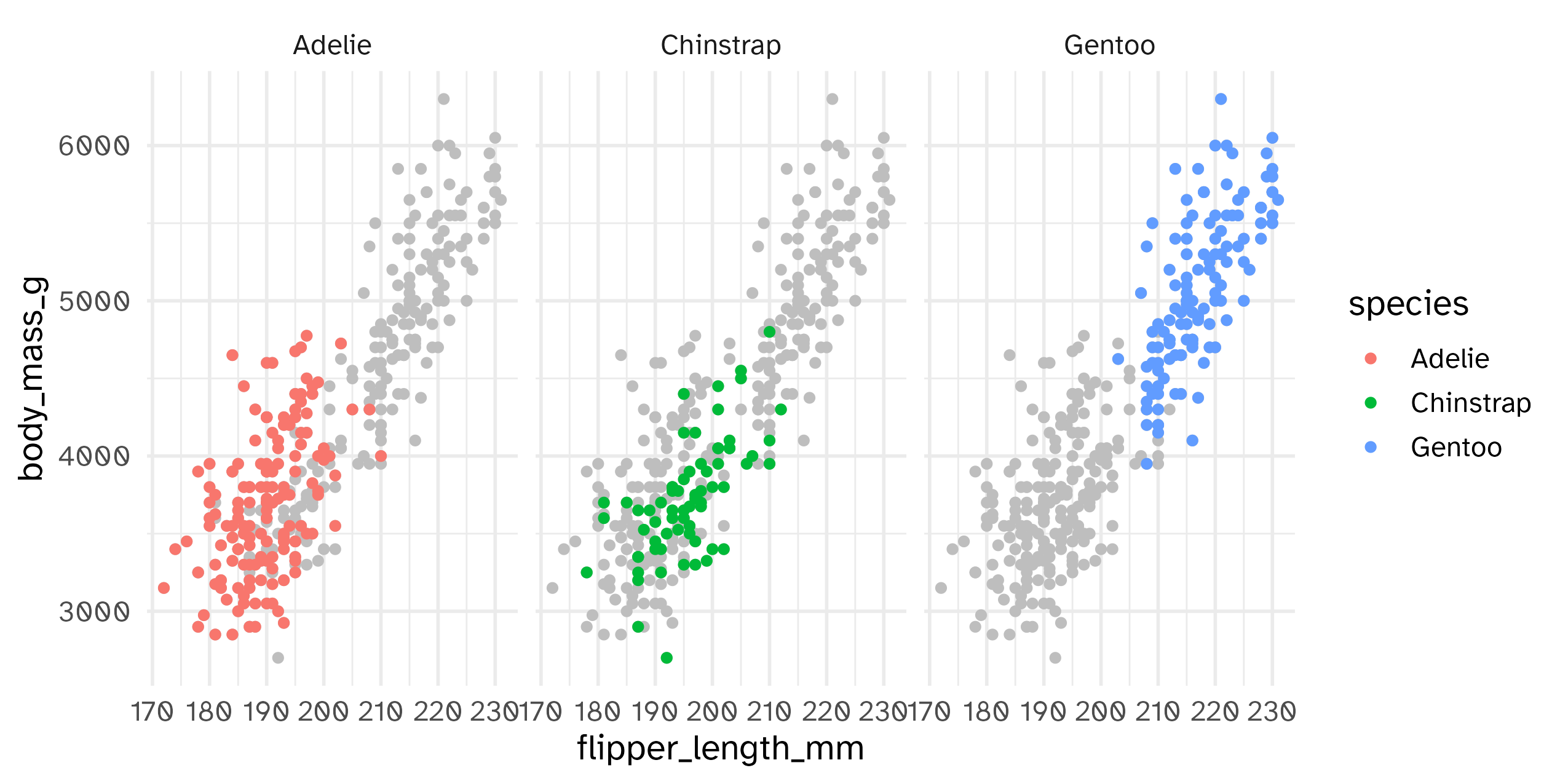
Themes
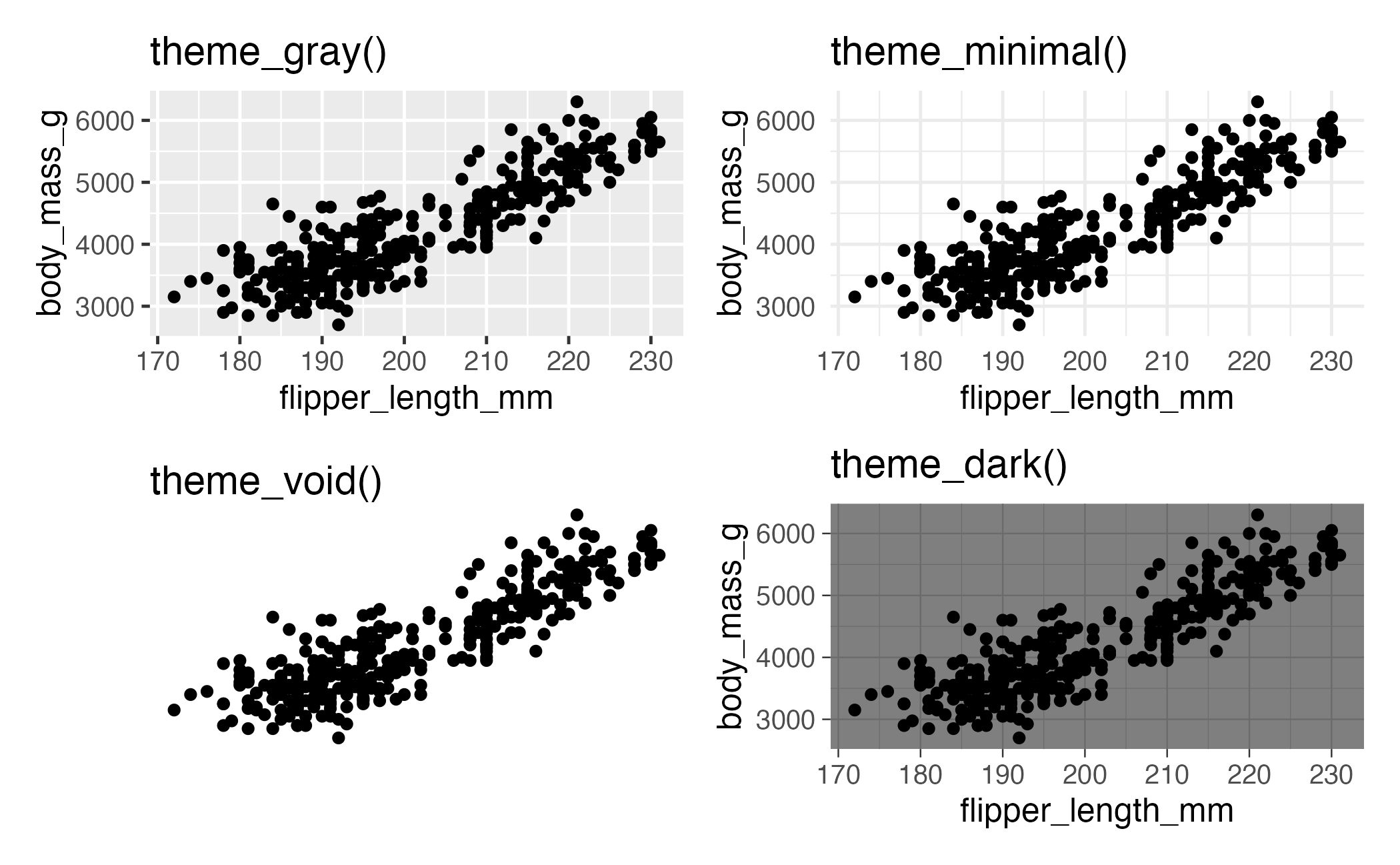
Complete themes
Themes from {ggthemes}
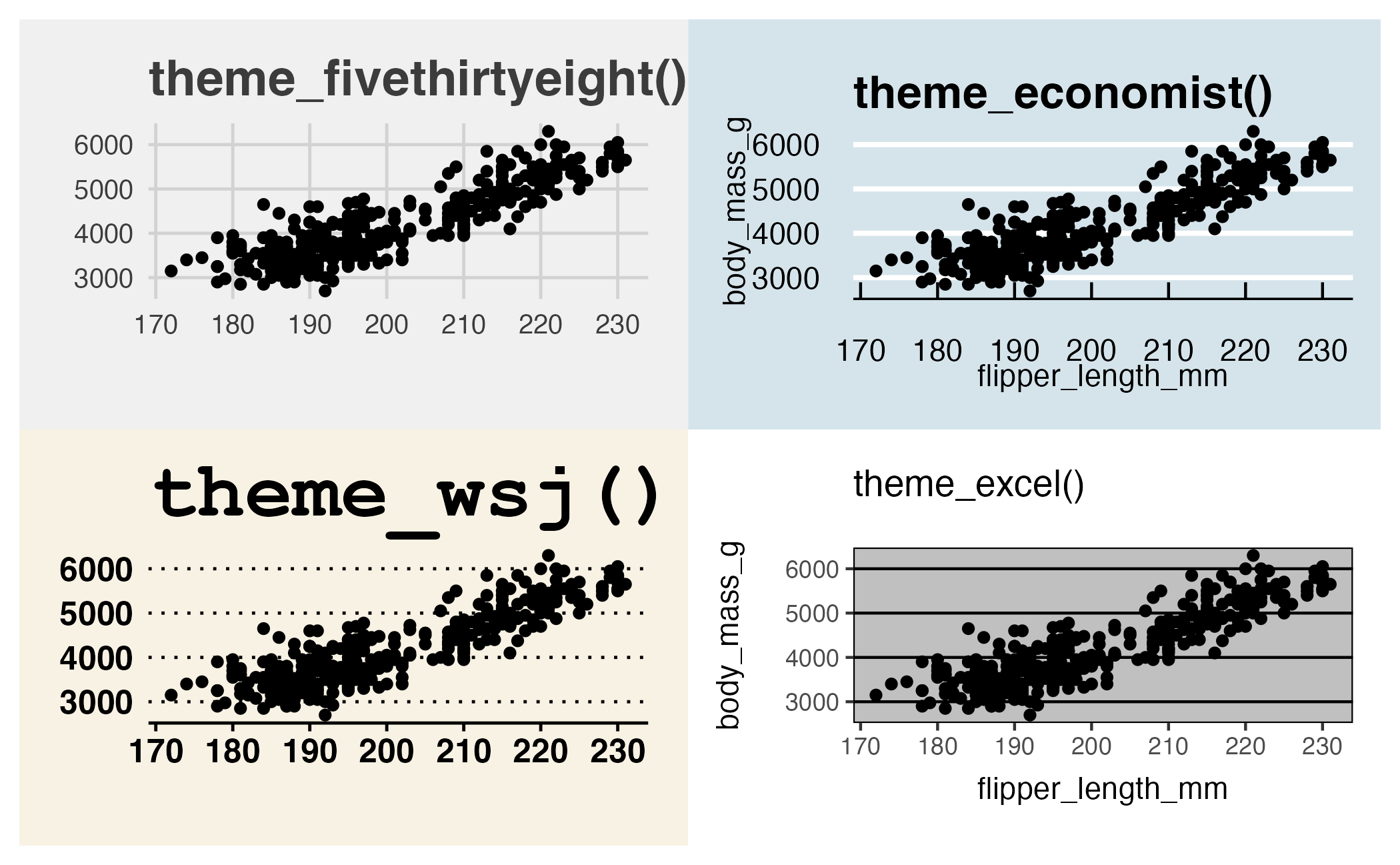
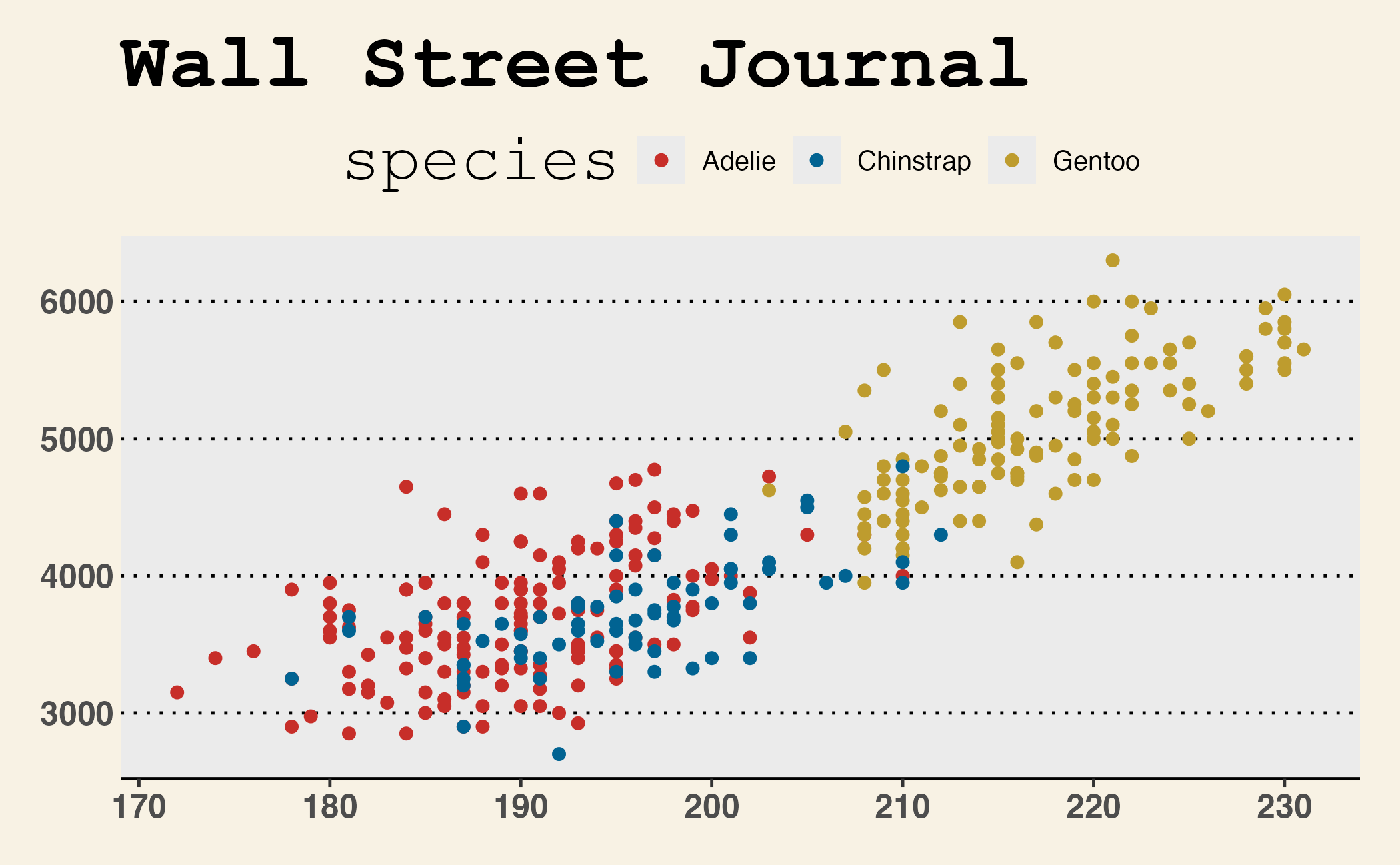
Themes and color scales from {ggthemes}
Modifying theme elements
Project 01
Project 01
- Initial proposal
- Develop as a team
- Take chances, make mistakes, get messy!

Wrap up
Recap
- Coordinate systems define how position aesthetics are drawn on the plot
- Limits and transformations work differently when applied to scales vs. coordinate systems
- Waffle charts are an alternative to pie charts for visualizing proportions
- Faceting generates small multiple window plots
Acknowledgements
- Slides derived in part from STA 313: Advanced Data Visualization