# A tibble: 51 × 4
long lat survivors direction
<dbl> <dbl> <dbl> <chr>
1 24 54.9 340000 A
2 24.5 55 340000 A
3 25.5 54.5 340000 A
4 26 54.7 320000 A
5 27 54.8 300000 A
6 28 54.9 280000 A
7 28.5 55 240000 A
8 29 55.1 210000 A
9 30 55.2 180000 A
10 30.3 55.3 175000 A
# ℹ 41 more rowsThe grammar of graphics
Lecture 2
Dr. Benjamin Soltoff
Cornell University
INFO 3312/5312 - Spring 2026
January 22, 2026
Announcements
Announcements
- Homework 01 released this evening and due next week
- Waitlist update
- 7 pins distributed so far
- INFO 3312: 0 seats available and 28 on the waitlist (14 IS majors)
- INFO 5312: 0 seats available and 7 on the waitlist
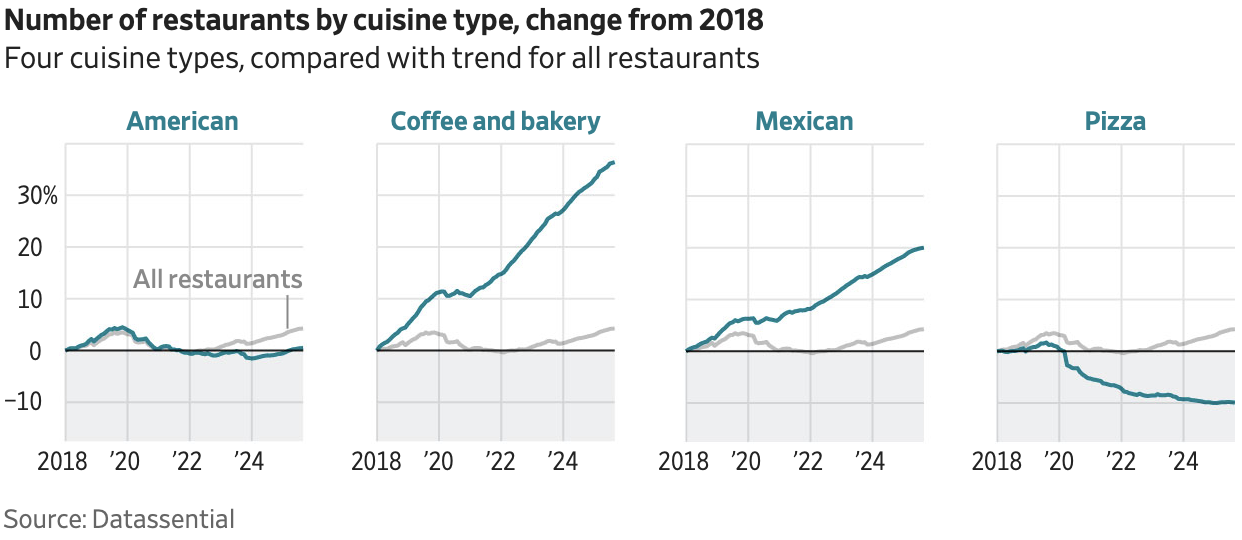
Number of restaurants by cuisine type
Source: The Wall Street Journal
Learning objectives
- Utilize the grammar of graphics to conceptually define Minard’s graph of Napoleon’s invasion of Russia
- Map variables to aesthetics
- Create small-multiples plots using faceting
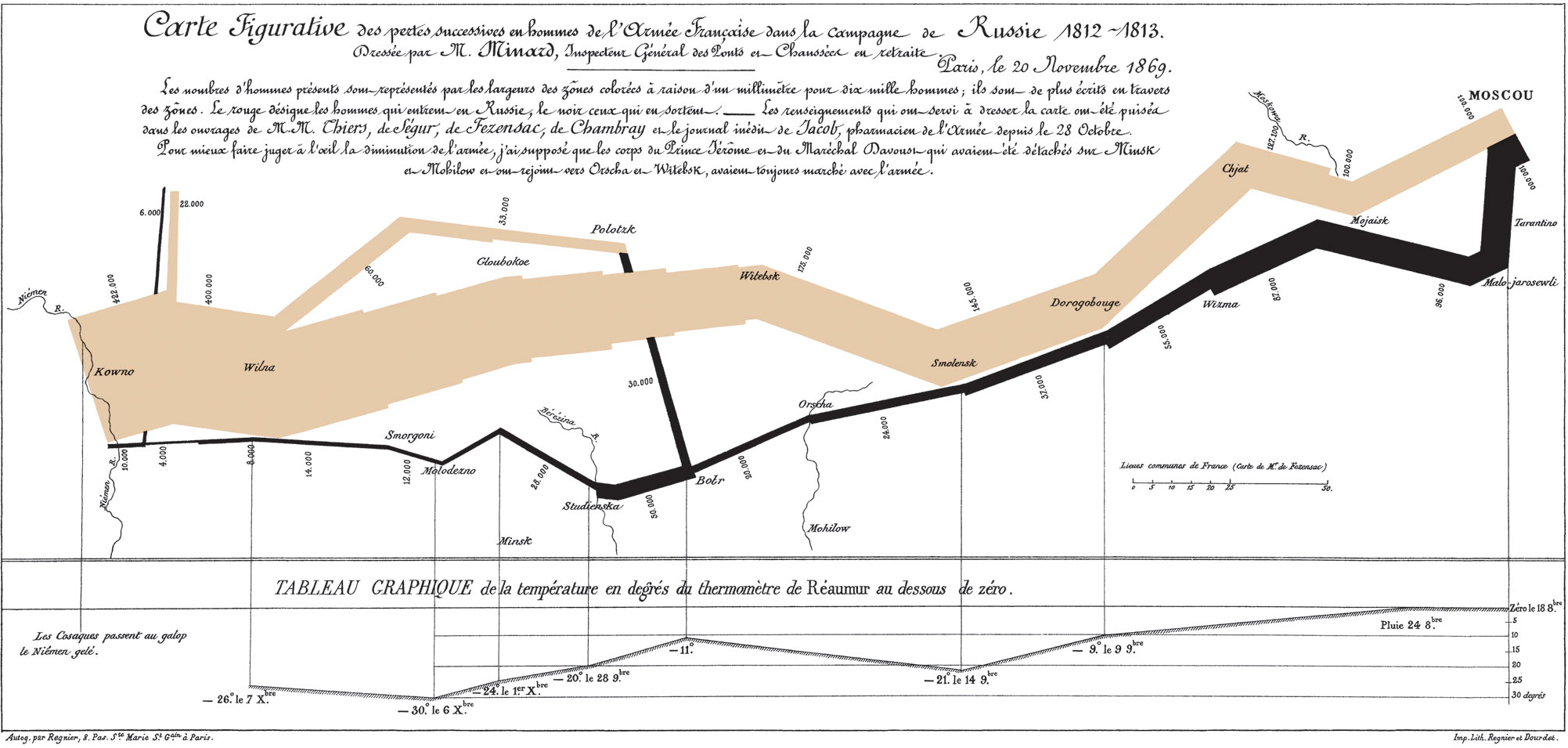
“The best statistical graphic ever drawn”
Source: Wikipedia
Building Minard’s map in R
troops
cities
# A tibble: 20 × 3
long lat city
<dbl> <dbl> <chr>
1 24 55 Kowno
2 25.3 54.7 Wilna
3 26.4 54.4 Smorgoni
4 26.8 54.3 Moiodexno
5 27.7 55.2 Gloubokoe
6 27.6 53.9 Minsk
7 28.5 54.3 Studienska
8 28.7 55.5 Polotzk
9 29.2 54.4 Bobr
10 30.2 55.3 Witebsk
11 30.4 54.5 Orscha
12 30.4 53.9 Mohilow
13 32 54.8 Smolensk
14 33.2 54.9 Dorogobouge
15 34.3 55.2 Wixma
16 34.4 55.5 Chjat
17 36 55.5 Mojaisk
18 37.6 55.8 Moscou
19 36.6 55.3 Tarantino
20 36.5 55 Malo-JarosewiiApplication exercise
ae-01
Aesthetics
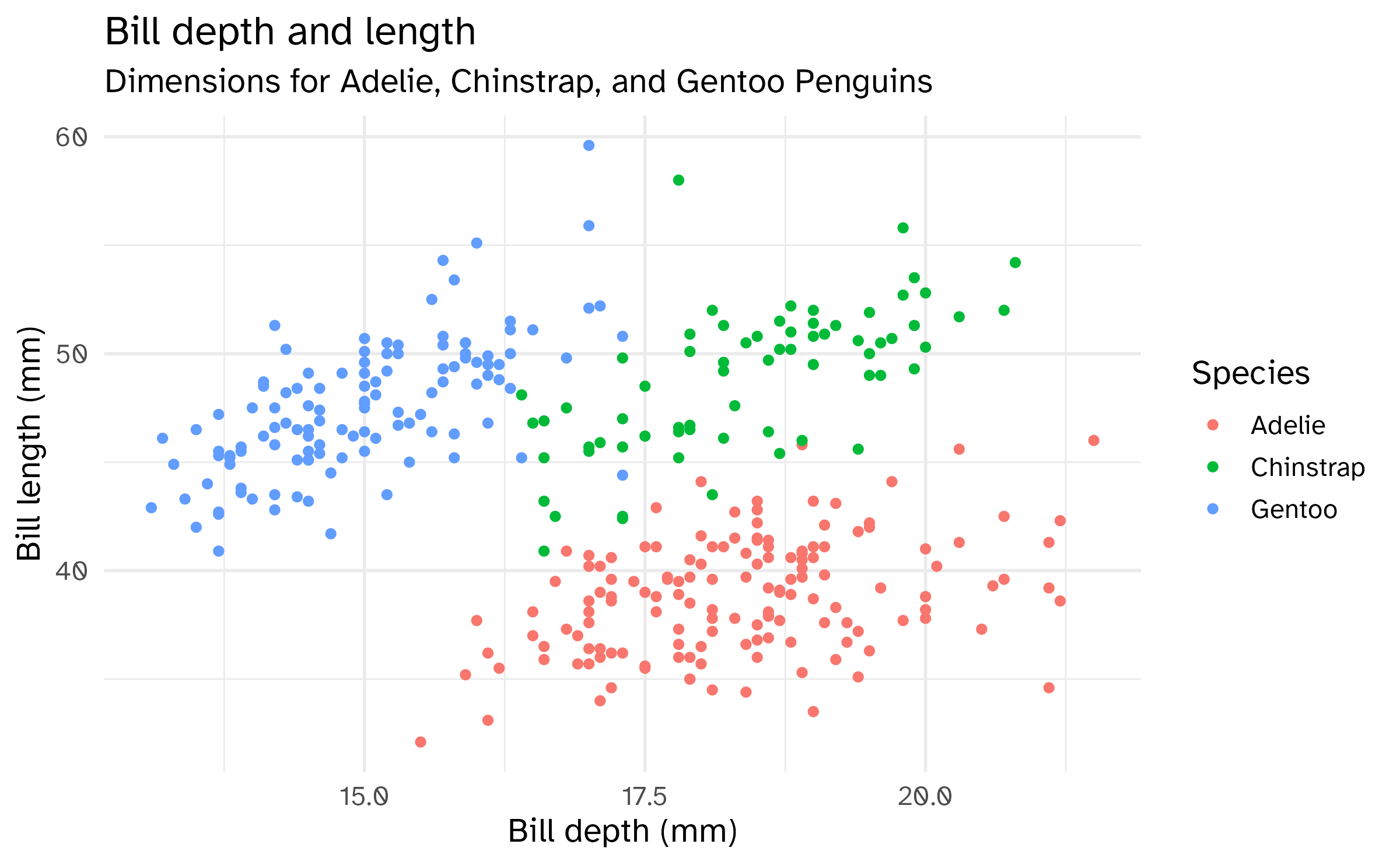
Aesthetics options
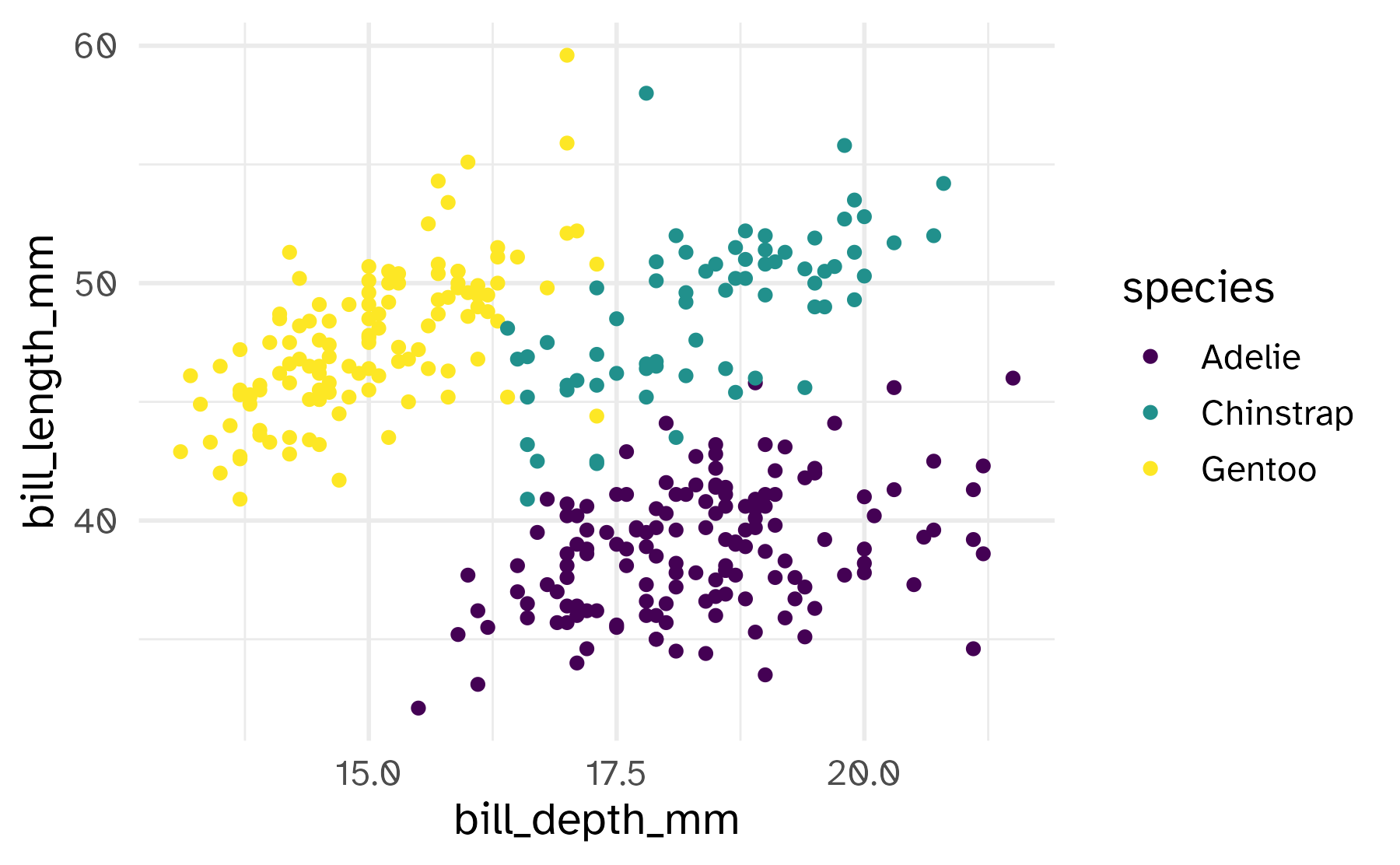
Commonly used characteristics of plotting characters that can be mapped to a specific variable in the data are
colorshapesizealpha(transparency)
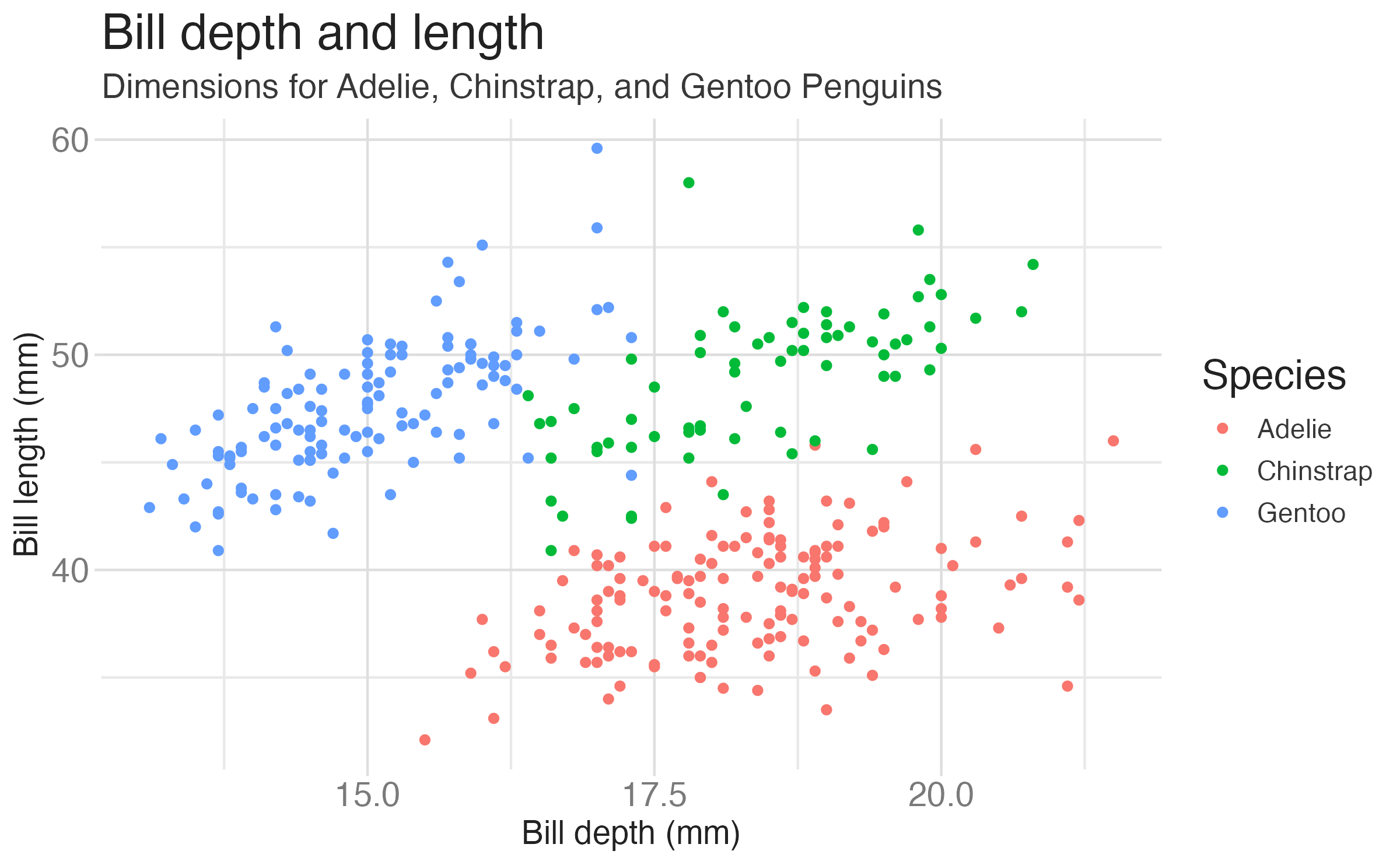
Color
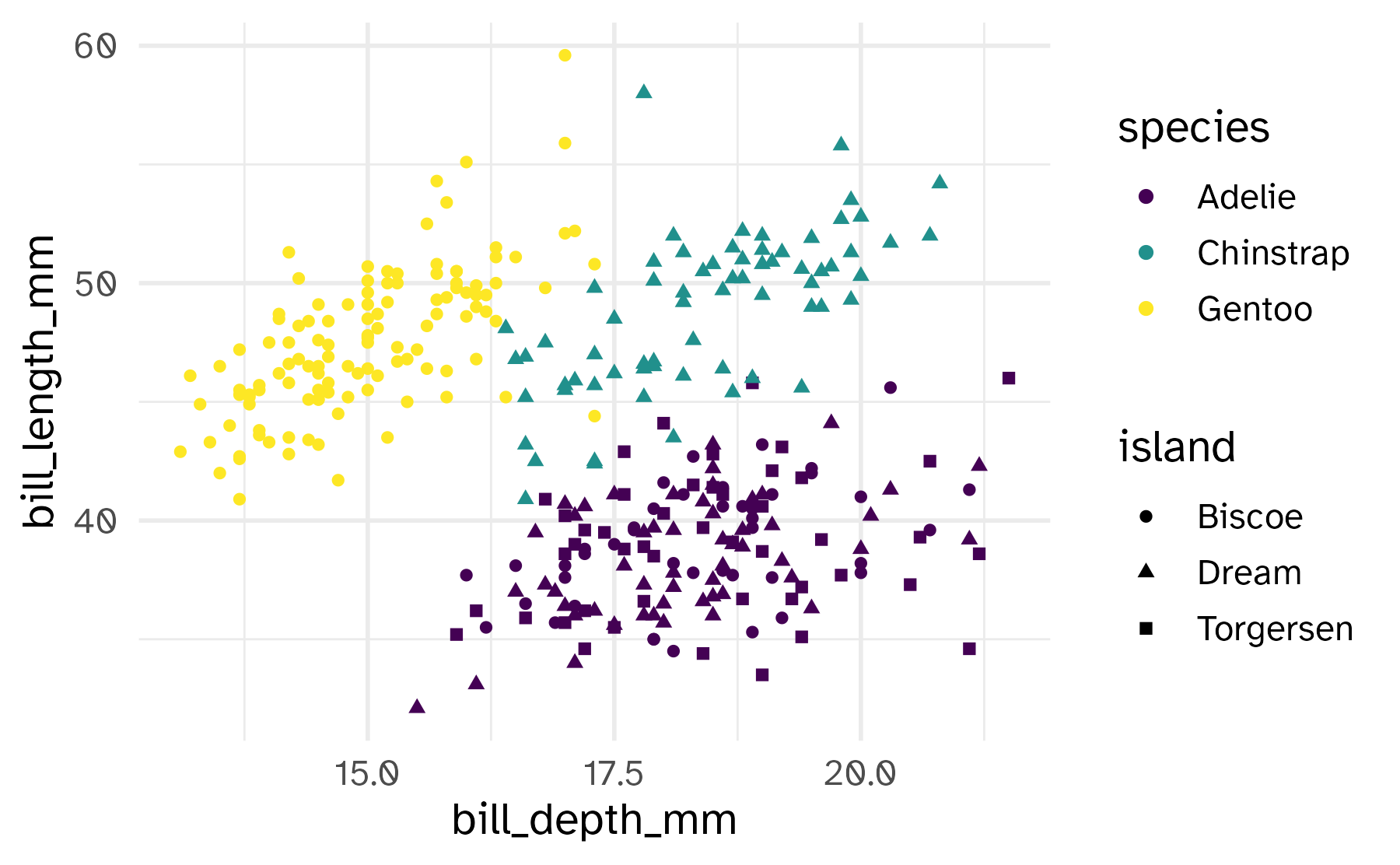
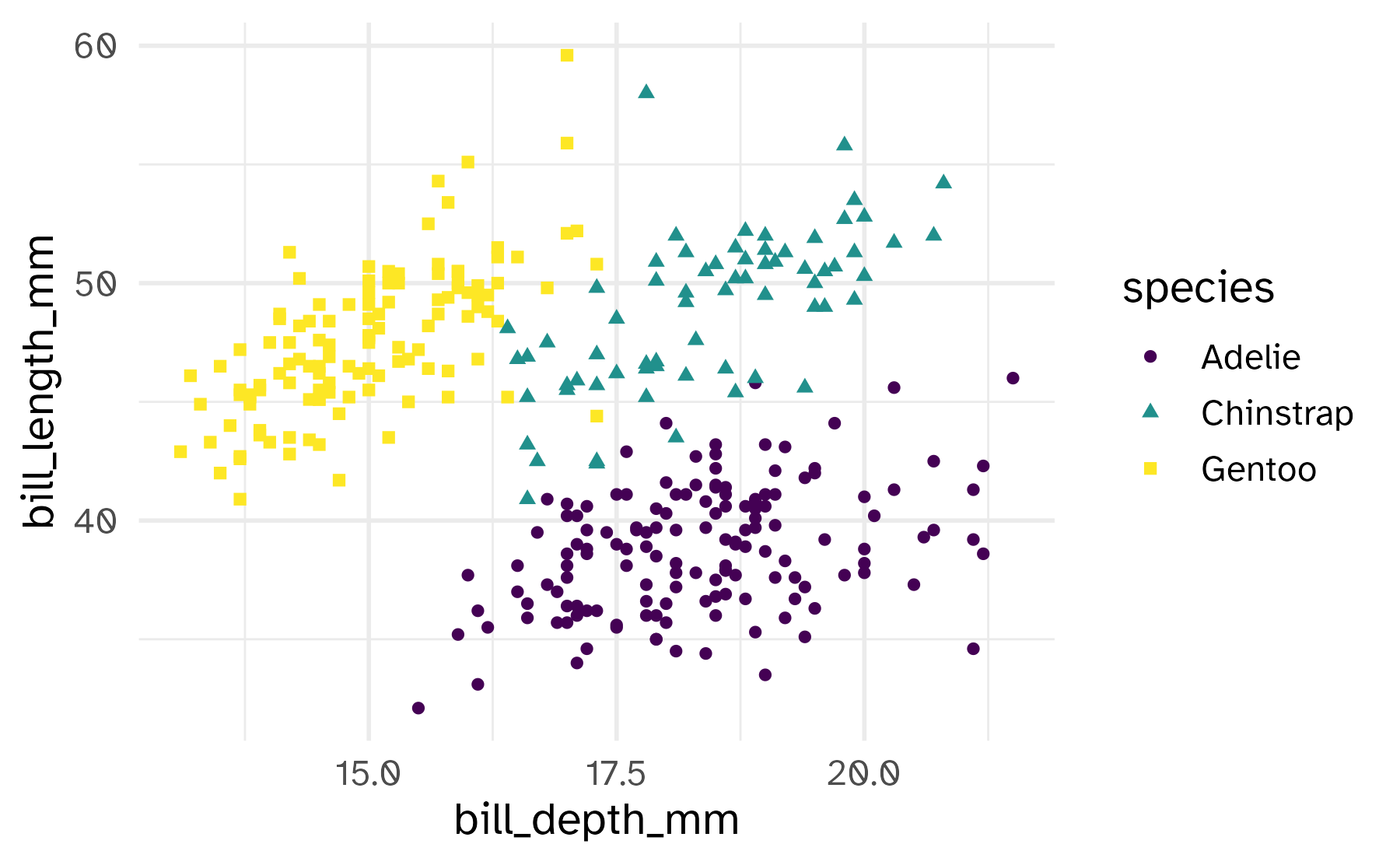
Shape
Mapped to a different variable than color
Shape
Mapped to same variable as color
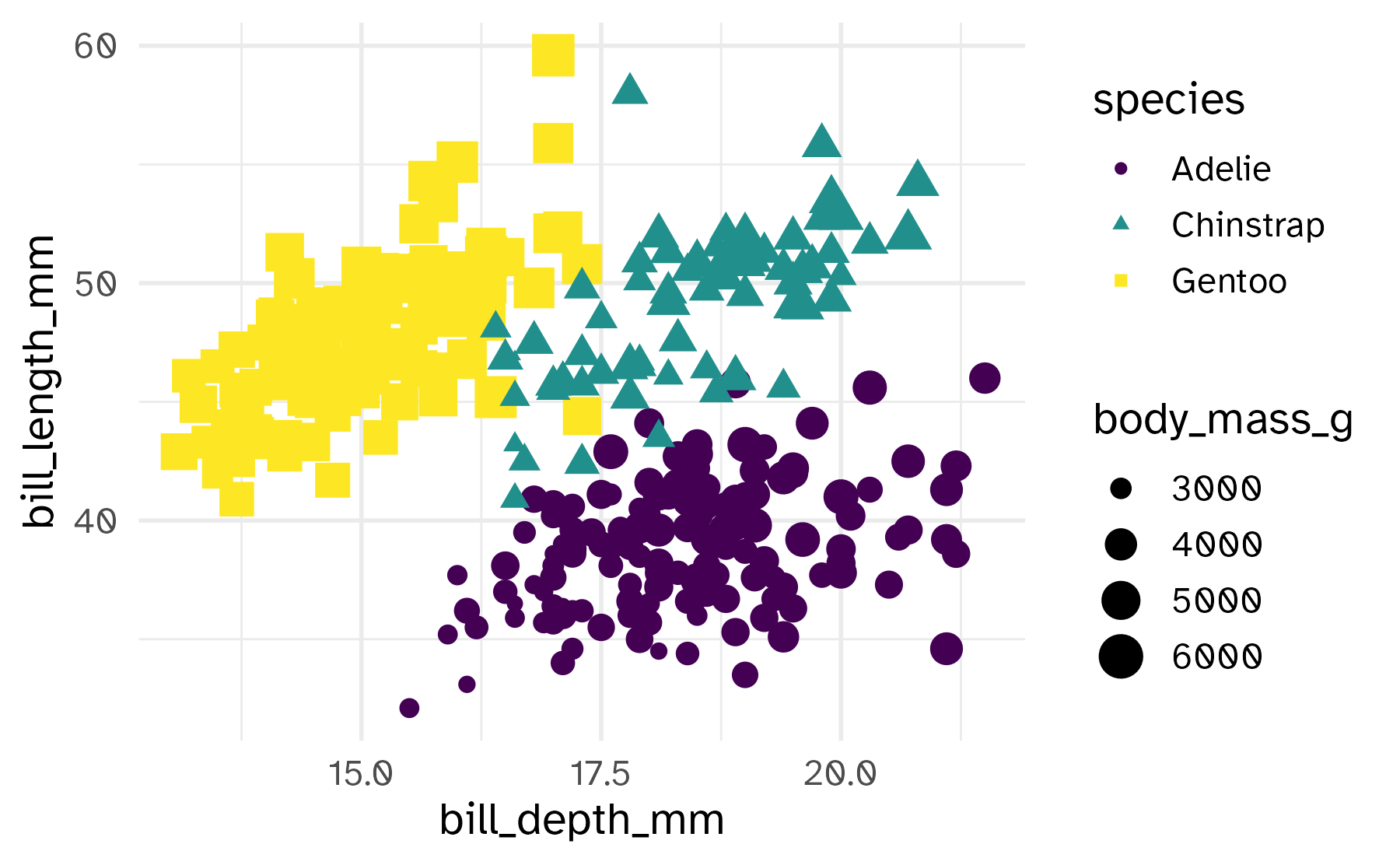
Size
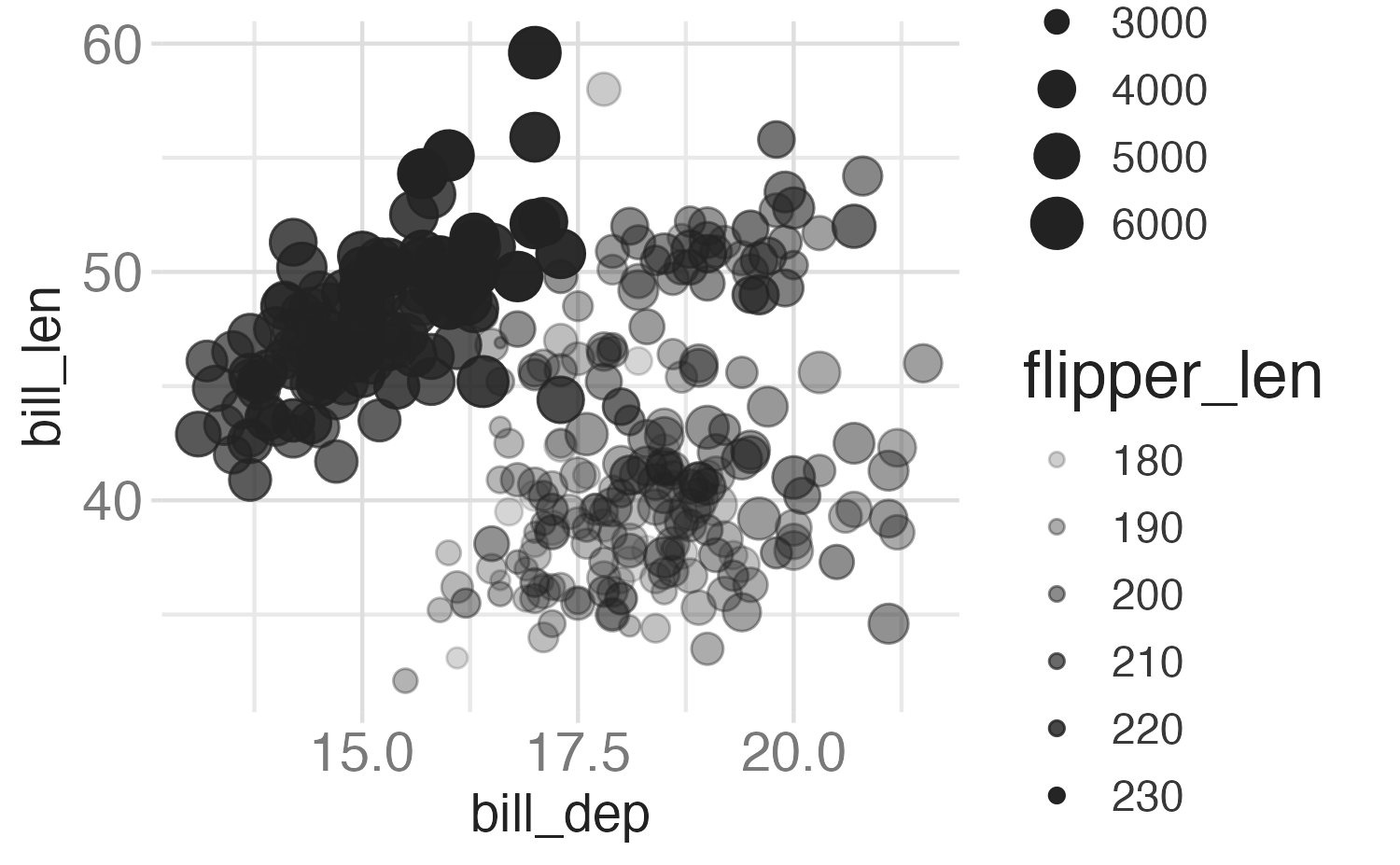
Alpha
Mapping vs. setting
- Mapping: Determine the size, alpha, etc. of points based on the values of a variable in the data
- goes into
aes()
- goes into
- Setting: Determine the size, alpha, etc. of points not based on the values of a variable in the data
- goes into
geom_*()
- goes into
Faceting
Faceting
- Smaller plots that display different subsets of the data
- Useful for exploring conditional relationships and large data
Various ways to facet
In the next few slides describe what each plot displays. Think about how the code relates to the output.
Note: The plots in the next few slides do not have proper titles, axis labels, etc. because we want you to figure out what’s happening in the plots. But you should always label your plots!
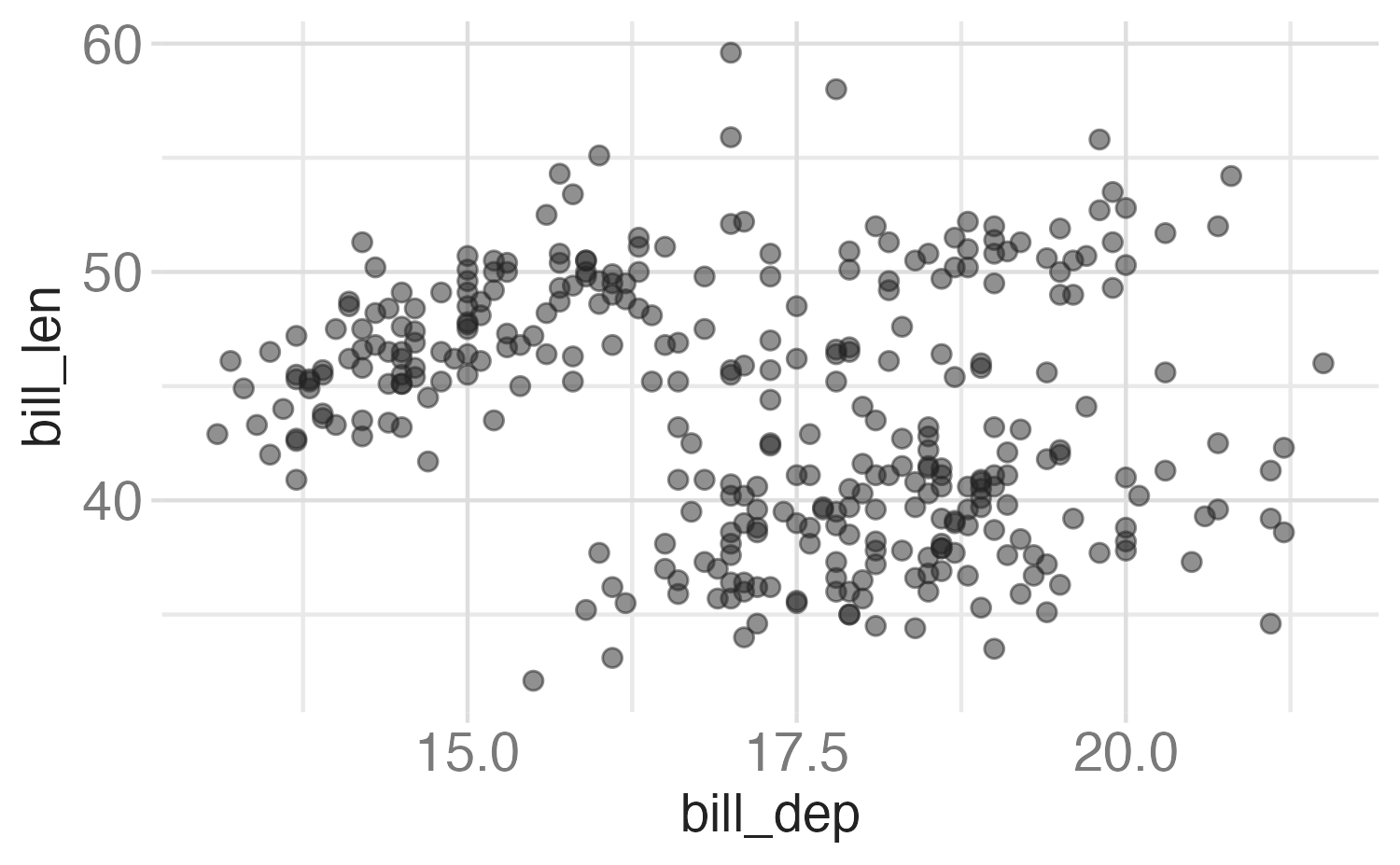
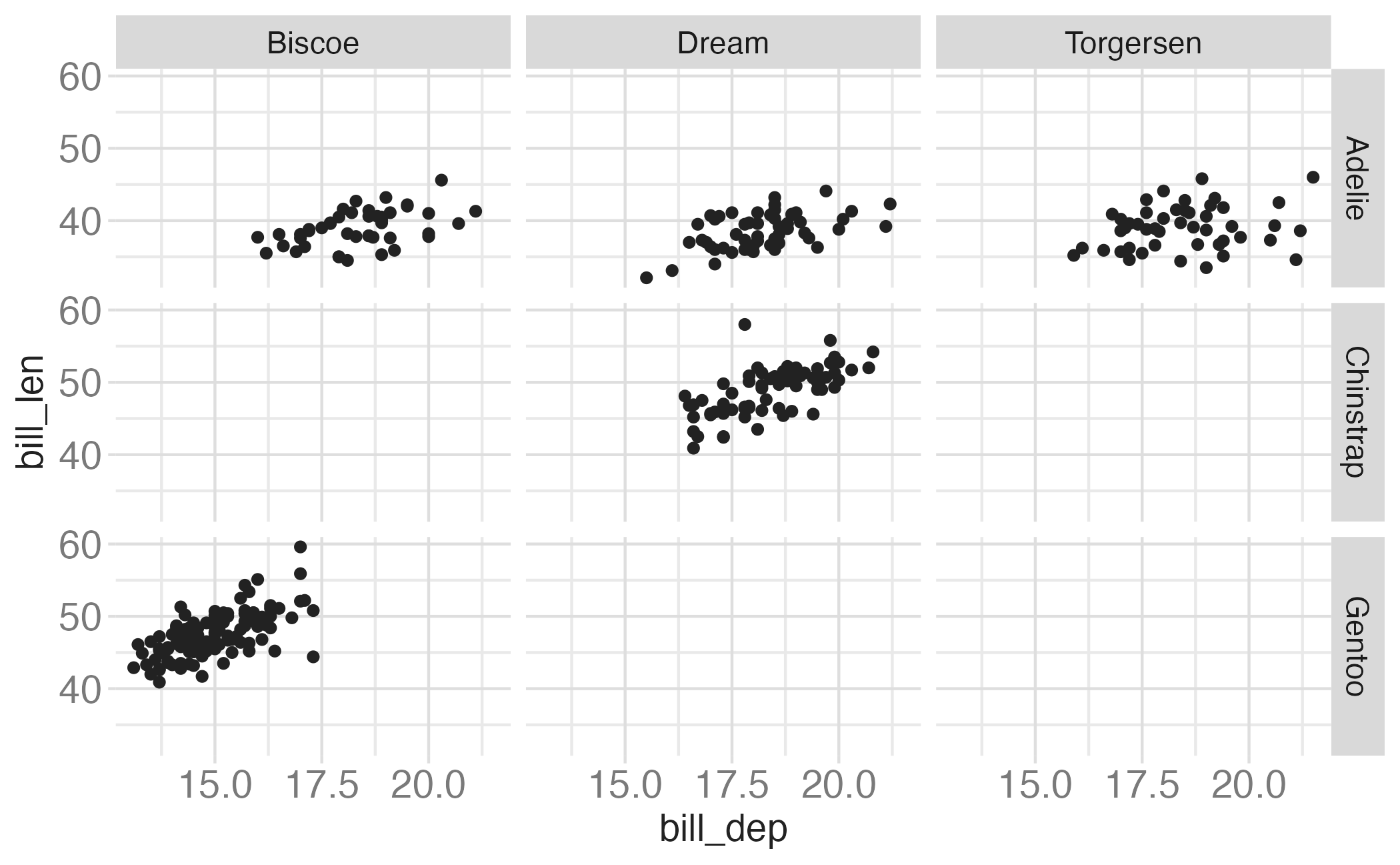
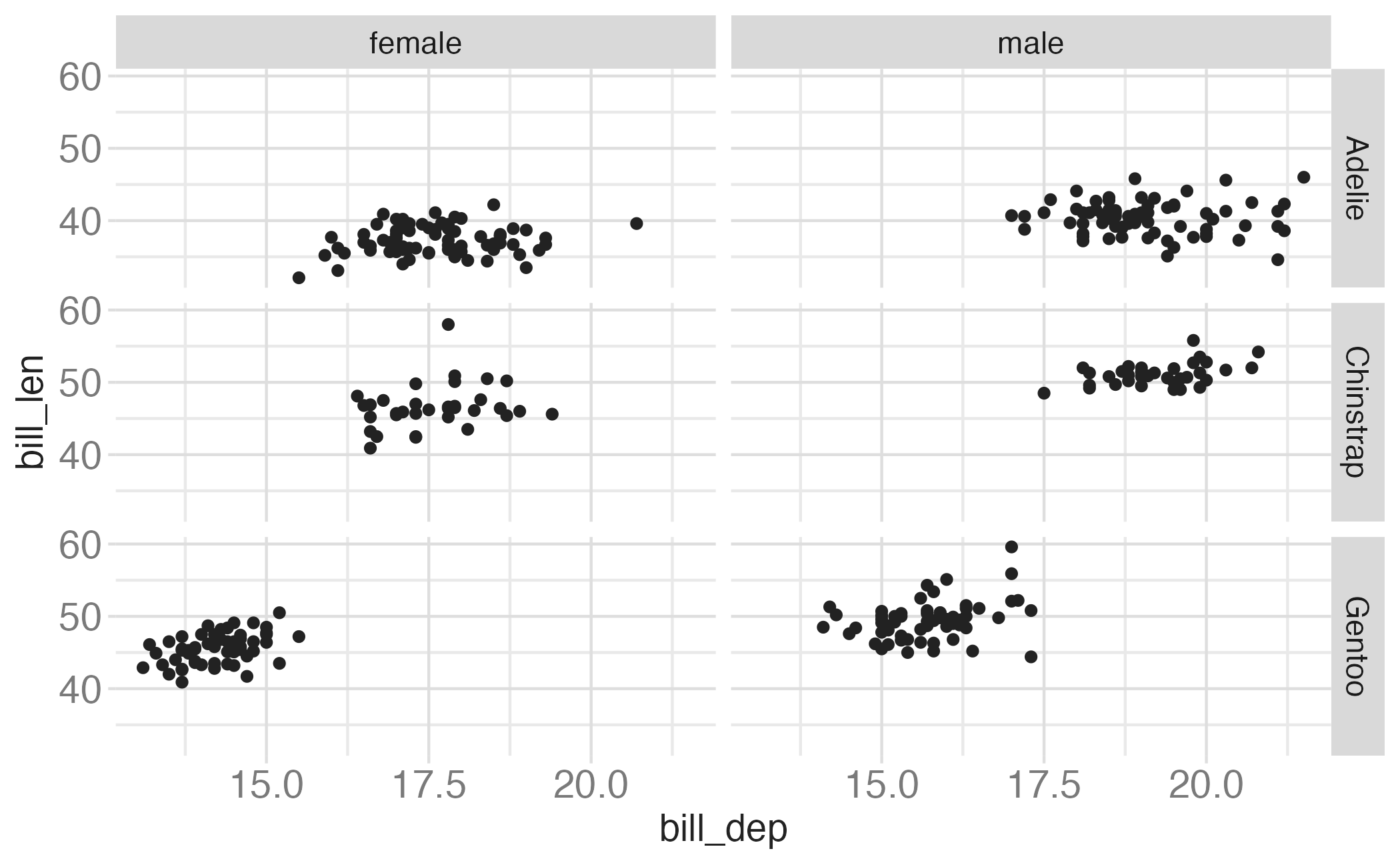
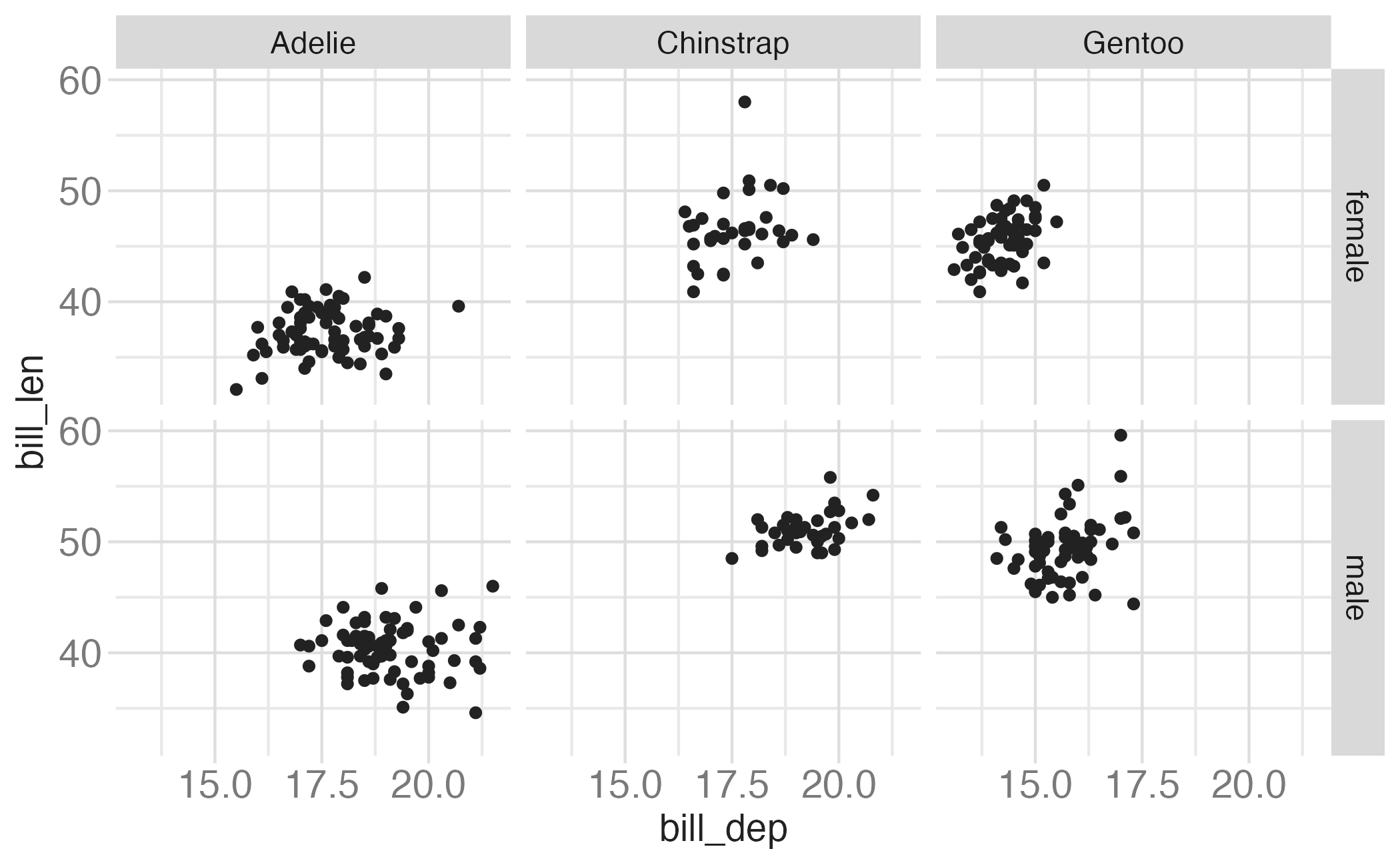
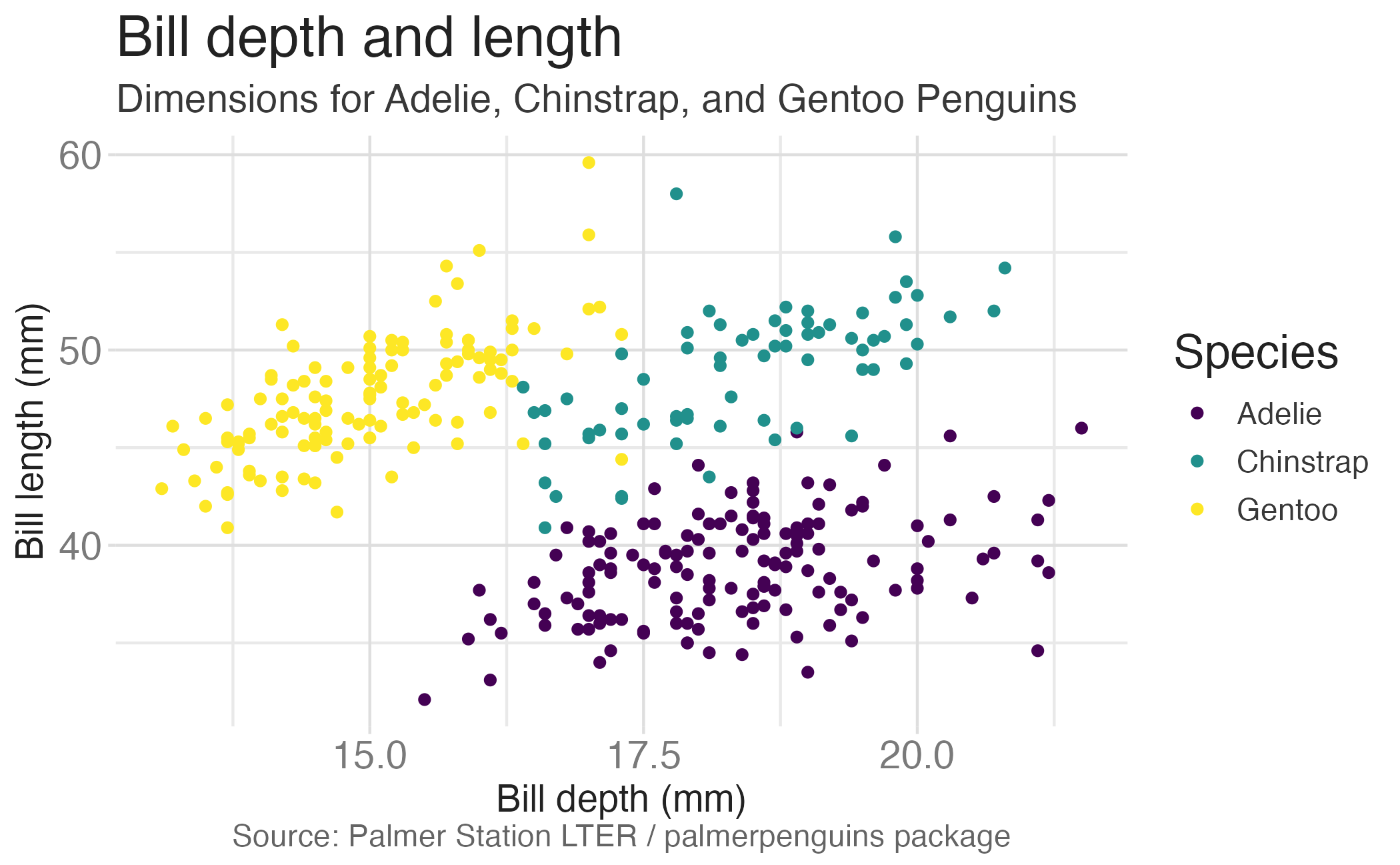
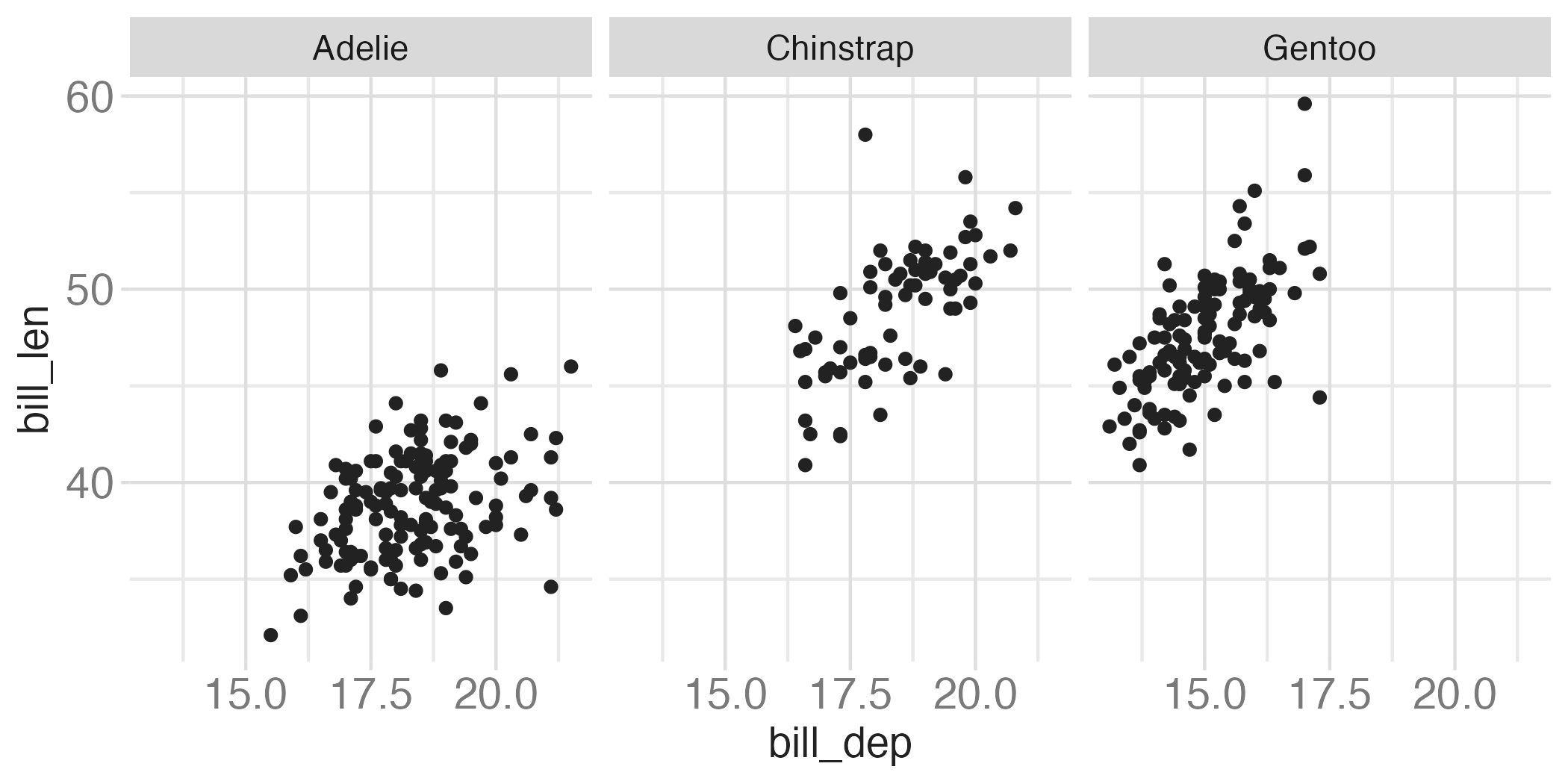
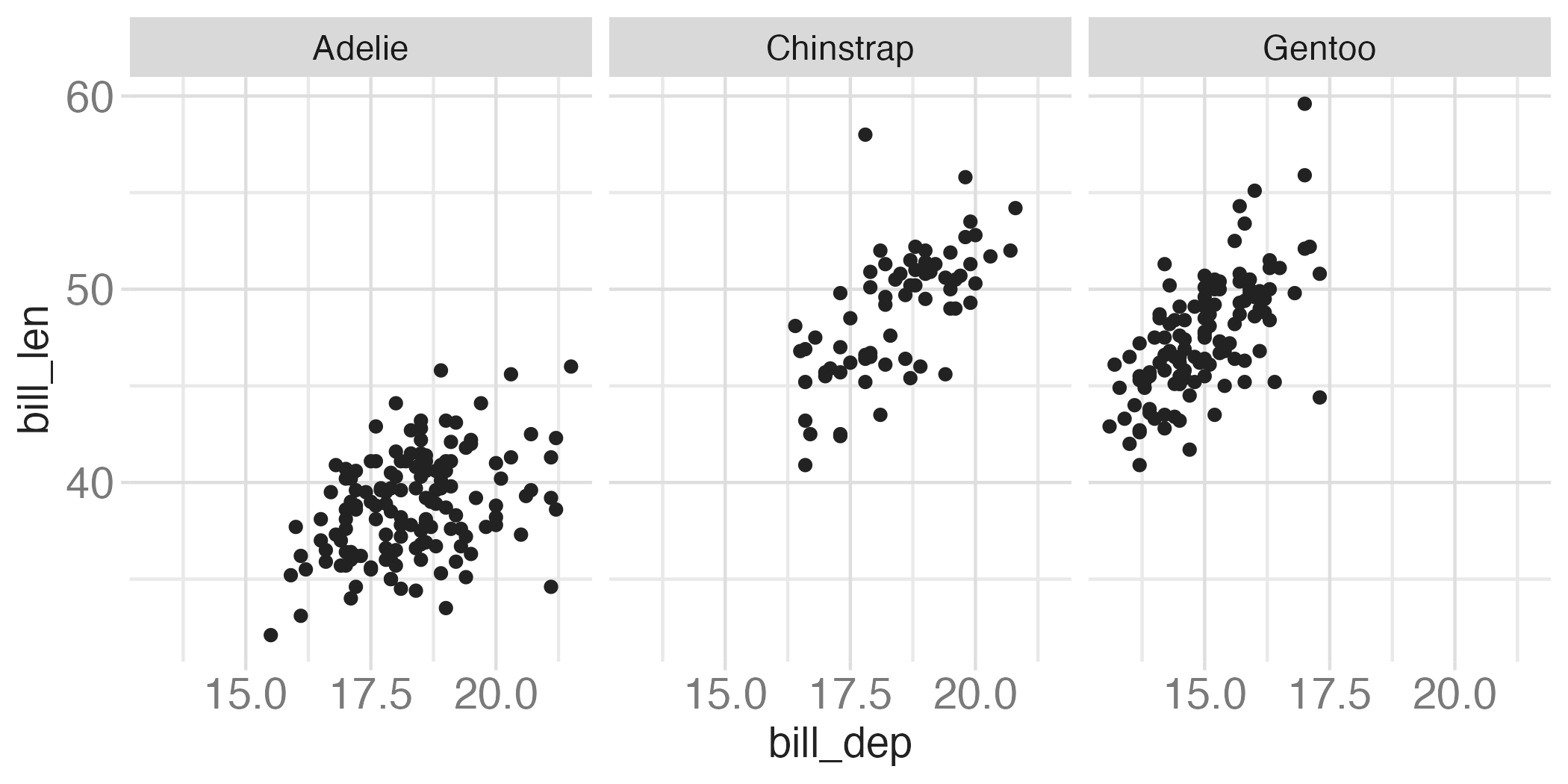
Faceting summary
facet_grid():- 2 dimensional grid
rows = vars(<VARIABLE>), cols = vars(<VARIABLE>)- Alternative:
rows ~ cols
facet_wrap(): 1 dimensional ribbon wrapped according to number of rows and columns specified or available plotting area
Facet and color
Facet and color, no legend
Wrap up
Recap
- {ggplot2} is based on the grammar of graphics
- Use the
ggplot()function to initialize a plot aes()maps variables to aesthetics- Use
geom_*()to add geoms to a plot - Use
facet_*()to facet a plot
Acknowledgements
- Slides derived in part from Data Science in a Box