Rows: 344
Columns: 8
$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie…
$ island <fct> Torgersen, Torgersen, Torgersen, Torge…
$ bill_len <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9…
$ bill_dep <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8…
$ flipper_len <int> 181, 186, 195, NA, 193, 190, 181, 195,…
$ body_mass <int> 3750, 3800, 3250, NA, 3450, 3650, 3625…
$ sex <fct> male, female, female, NA, female, male…
$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 20…Welcome to INFO 3312/5312
Lecture 1
Cornell University
INFO 3312/5312 - Spring 2026
January 20, 2026
Announcements
Learning objectives
- Introduce the course and its structure
- Identify the course learning objectives and how we will learn to visualize data
- Review course policies
- Discuss hopes and dreams for the course
- Introduce the grammar of graphics
Course details
Timetable
- Lectures
- Tuesdays 1:25-2:40pm
- Thursdays 1:25-2:40pm
- Course is 3 credits - no Friday lab section
Students on the waitlist
- INFO 3312/5312 enrollment is restricted to IS/ISST majors and IS MPS students
- If you are not an IS/ISST major (or are still in the process of affiliating), join the waitlist through Student Center
- PINs distributed on a rolling basis
- As of January 20 (12:10pm):
- INFO 3312: 0 seats available and 34 on the waitlist
- INFO 5312: 1 seats available and 6 on the waitlist
Themes: what, why, and how
- What: the communication (e.g. plot, table, report)
- Specific types of visualizations for a particular purpose (e.g., maps for spatial data, Sankey diagrams for proportions, etc.)
- Tooling to produce them (e.g., specific R packages)
- How: the process
- Start with a design (sketch + pseudo code)
- Pre-process data (e.g., wrangle, reshape, join, etc.)
- Map data to aesthetics
- Make visual encoding decisions (e.g., address accessibility concerns)
- Post-process for visual appeal and annotation
- Why: the theory
- Tie together “how” and “what” through the grammar of graphics
- Extend to underlying theory of cognition and information processing
Introductions
Meet the instructor
Dr. Benjamin Soltoff
Associate Teaching Professor in Information Science
CIS Building 284

Meet the course team
| Name | Role(s) | |
|---|---|---|
| Xinyue He | GTRS | |

|
Catherine (Tianhong) Yu | PhD TA |
| Celina Jang | Undergraduate TA | |
| Evan Wu | Undergraduate TA |
Meet each other!
Activity
Form a small group (3-4 individuals) with people sitting around you
First, introduce yourselves to each other:
- Your name - Prof/Dr. Soltoff
- Your major - Political science
The last movie you saw - People We Meet on Vacation
- What you hope to get out of this class - a job?
Start with bad graphs – Share your examples of “bad” graphs and why you think they’re bad.
Then, share good graphs – Same deal, share your examples of “good” graphs and why you think they’re good.
Finally, share your graphs on Canvas as comments on this discussion post.
Course components
Homepage
https://info3312.infosci.cornell.edu/
- All course materials
- Links to Canvas, GitHub, Posit Workbench, etc.
- Let’s take a tour!
Course toolkit
All linked from the course website:
GitHub organization: github.coecis.cornell.edu/info3312-sp26
Positron
Use the Workbench: posit-workbench.infosci.cornell.edu
Communication: GitHub Discussions
Assignment submission and feedback: Gradescope
Important
Make sure you can access Positron before Friday.
Activities: Prepare, Participate, Practice, Perform
Prepare: Introduce new content and prepare for lectures by completing the readings
Participate: Attend and actively participate in lectures, office hours, team meetings
Practice: Practice applying visualization techniques with application exercises during lecture, graded for completion
Perform: Put together what you’ve learned to analyze real-world data
- Homework assignments x 6-ish (individual)
- Team projects (2)
- Mini-project
Teams
- Team assignments
- Assigned by course staff
- Peer evaluation after completion
- Expectations and roles
- Everyone is expected to contribute equal effort
- Everyone is expected to understand all code turned in
- Individual contribution evaluated by peer evaluation, commits, etc.
Grading
| Category | Percentage |
|---|---|
| Project 1 | 20% |
| Project 2 | 30% |
| Mini-project | 20% |
| Homework | 20% |
| Application exercises | 10% |
See course syllabus for how the final letter grade will be determined.
INFO 5312
Additional expectations:
- INFO 5312 homework will at times be graded against a more stringent rubric
- INFO 5312 students will be grouped together for all projects
15 minute rule
Support
- Attend office hours
- Ask and answer questions on the discussion forum
- Reserve email for questions on personal matters and/or grades
- Read the course support page
Diversity + inclusion
- I want you to feel like you belong in this class and are respected
- We are committed to full inclusion in education for all persons
- If you feel that we have failed these goals, please either let us know or report it, and we will address the issue
Accessibility
I want this course to be accessible to students with all abilities. Please feel free to let me know if there are circumstances affecting your ability to participate in class.
Course policies
As long as you meet
the prereqs
Source: Cornell University
Prerequisites
- INFO 2950/2951 or INFO 5001
- Prior experience with R and Git is required
Ideally you took INFO 2950/2951 or 5001 with me.
If not, you need a firm understanding of R (including {tidyverse}) and Git workflows.
Late work, waivers, regrades policy
- We have policies!
- Read about them on the course syllabus and refer back to them when you need it
Collaboration policy
Only work that is clearly assigned as team work should be completed collaboratively.
Homeworks must be completed individually. You may not directly share answers / code with others, however you are welcome to discuss the problems in general and ask for advice.
Sharing / reusing code policy
We are aware that a huge volume of code is available on the web, and many tasks may have solutions posted
Any recycled code that is discovered and is not explicitly cited will be treated as plagiarism, regardless of source.
All code must be written by you, the human being.
Generative AI
Use generative AI to facilitate, rather than hinder, learning
✅ GAI tools for reference purposes
✅ GAI tools for writing my code
You may use GAI tools to assist in writing code in this class
You may not make use of the technology as a substitute for critical thinking
I reserve the right to orally assess any student on their submissions to verify they meet the learning objectives for the assignment
❌ GAI tools for narrative
You are ultimately responsible for the work you turn in; it should reflect your understanding of the course content
Academic integrity
- A student shall in no way misrepresent his or her work.
- A student shall in no way fraudulently or unfairly advance his or her academic position.
- A student shall refuse to be a party to another student’s failure to maintain academic integrity.
- A student shall not in any other manner violate the principle of academic integrity.
Most importantly!
Ask if you’re not sure if something violates a policy!
The grammar of graphics
The grammar of graphics
- “The fundamental principles or rules of an art or science”
- A grammar used to describe and create a wide range of statistical graphics
- Originated by Leland Wilkinson in 2001, expanded by Hadley Wickham in 2005 with {ggplot2}

Image credit: @allison_horst
{ggplot2} \(\in\) {tidyverse}
Data: Palmer Penguins
Measurements for penguin species, island in Palmer Archipelago, size (flipper length, body mass, bill dimensions), and sex.
Example drawn from STA 313
Coding out loud
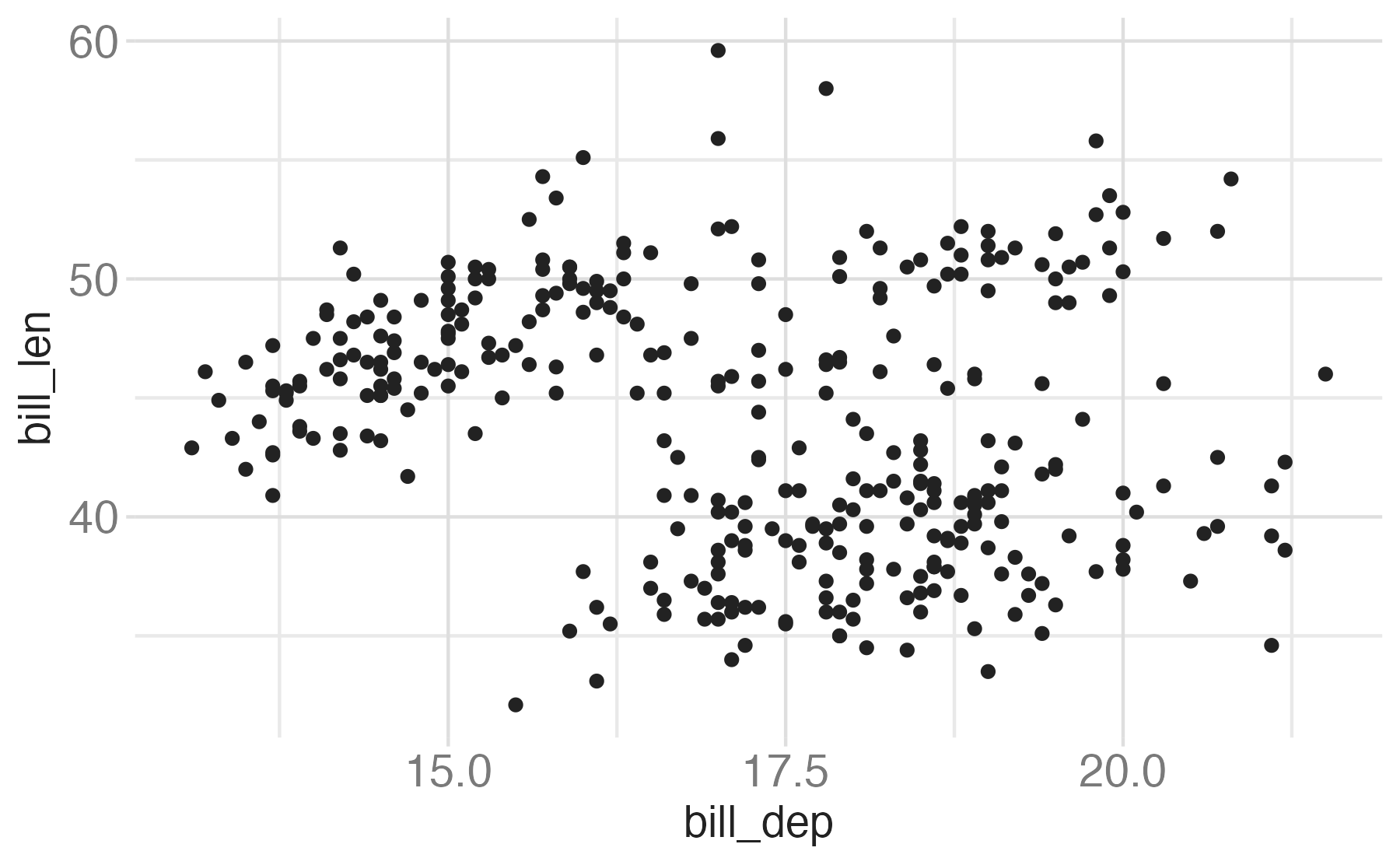
Start with the
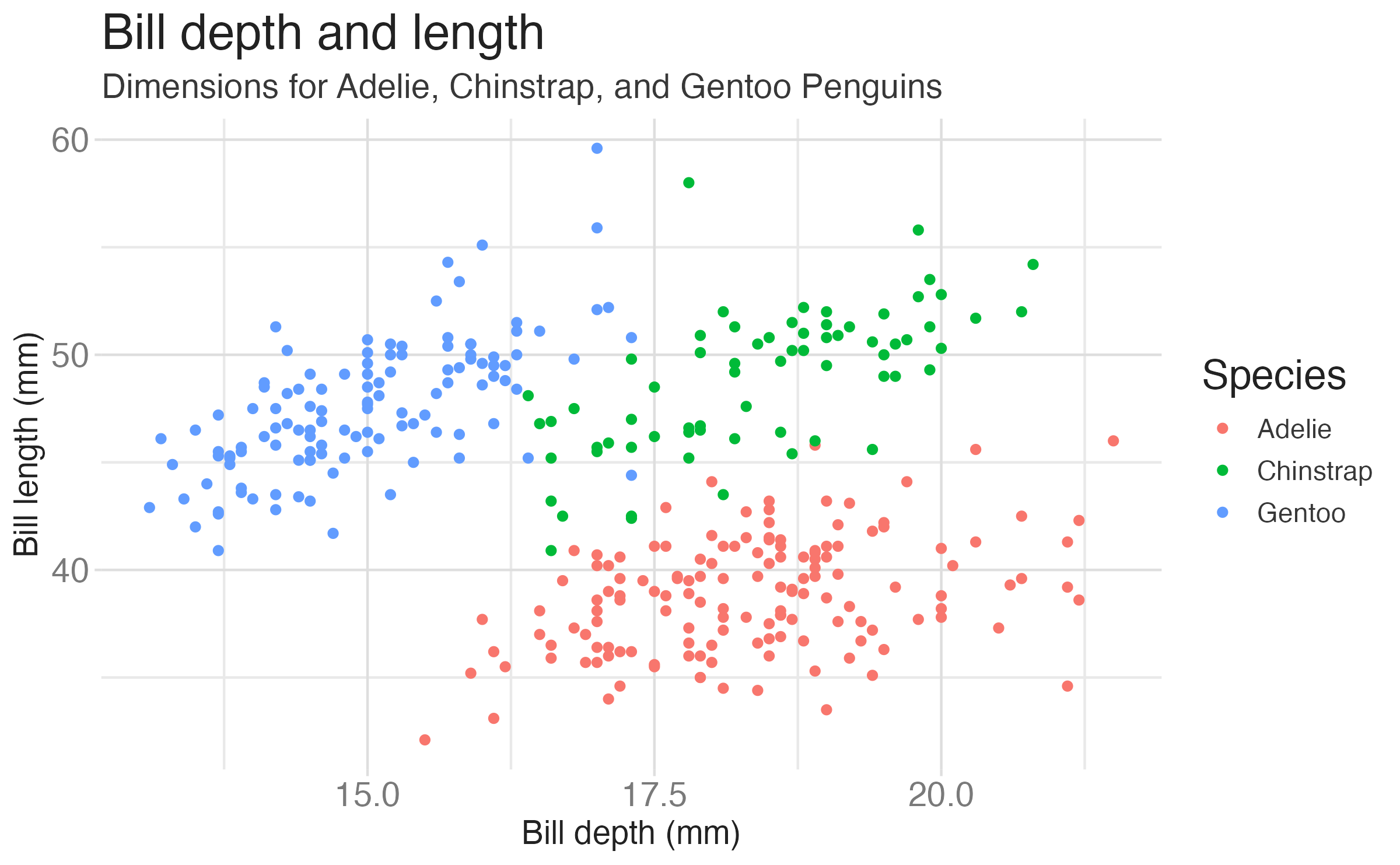
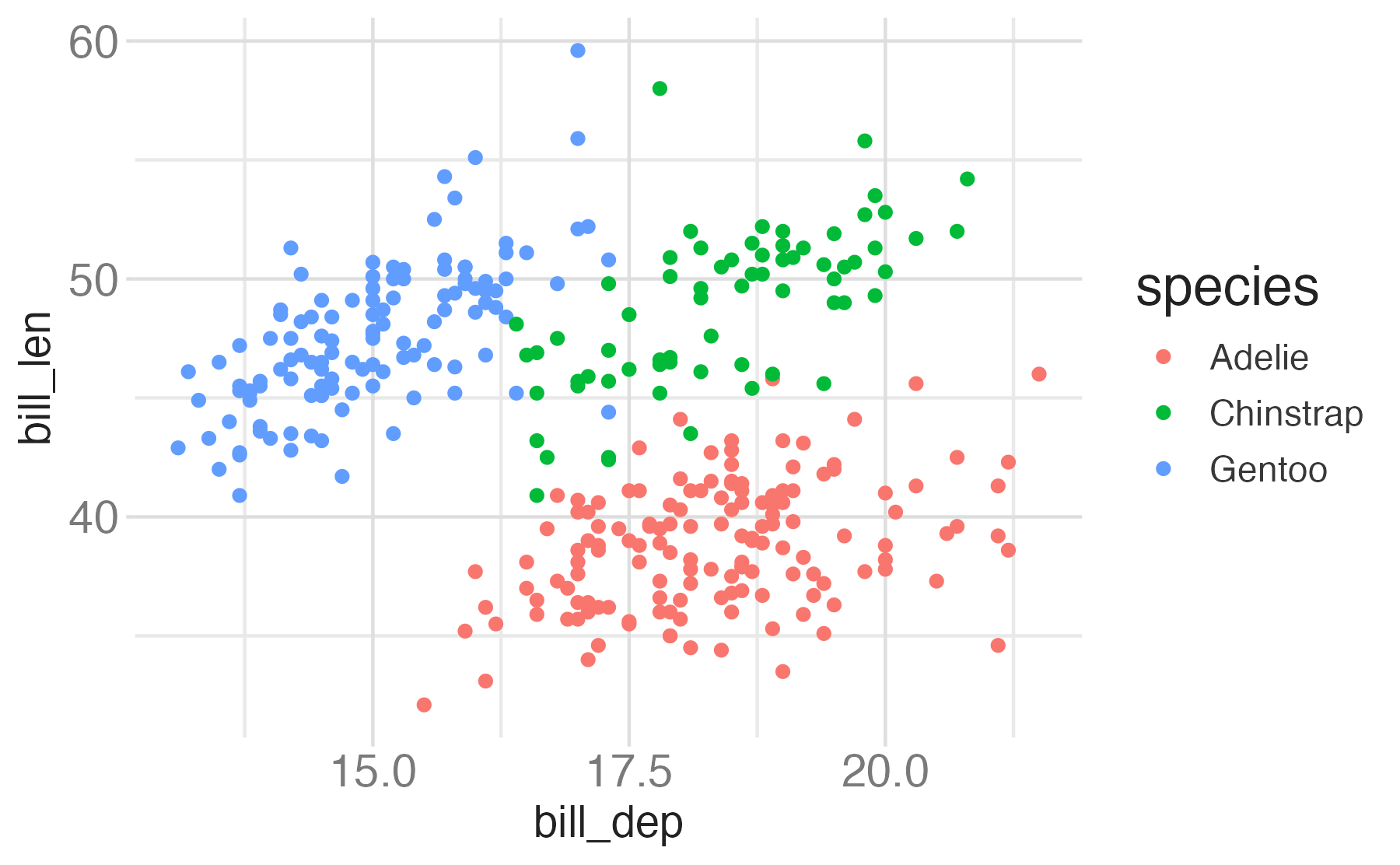
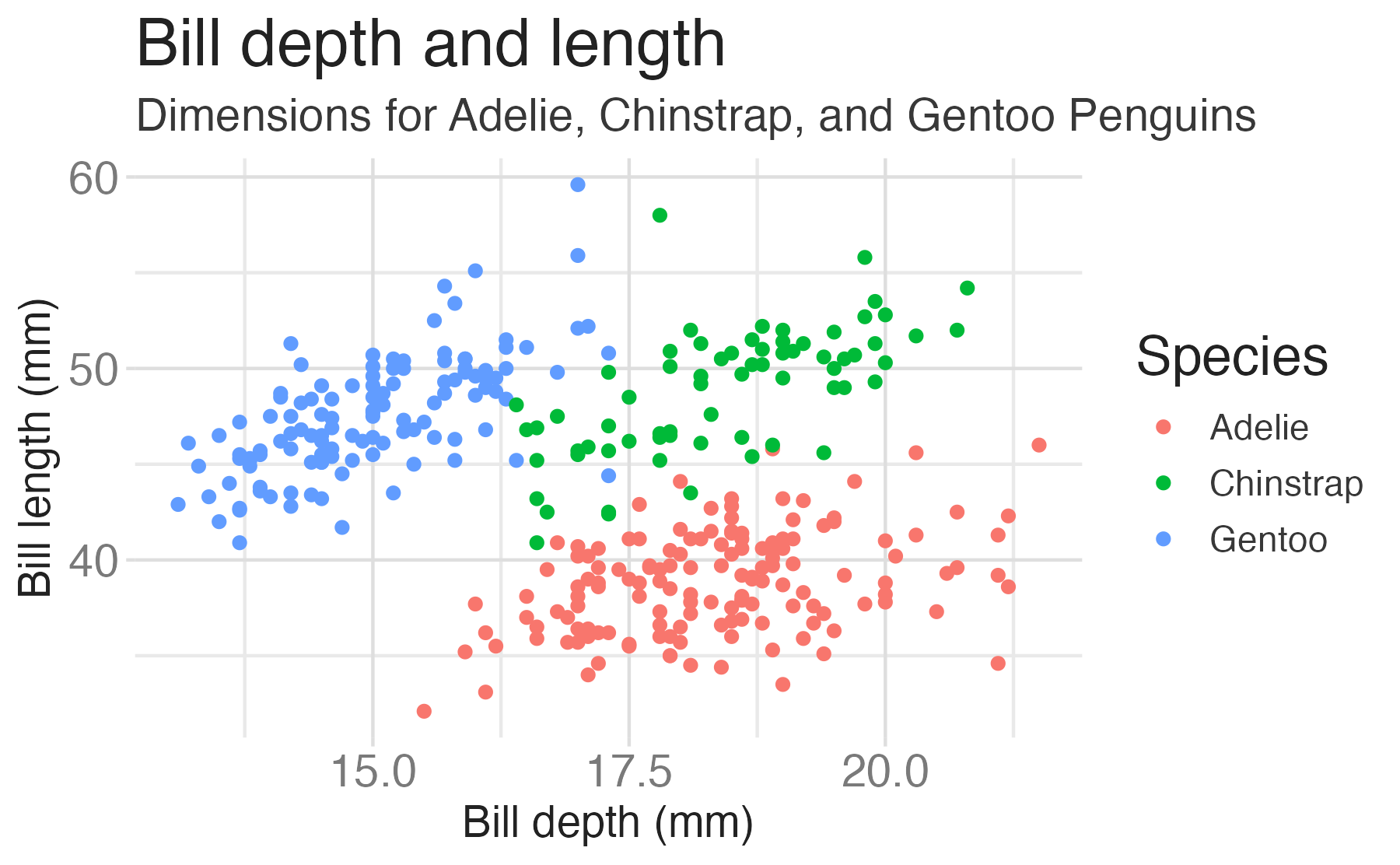
penguinsdata frame, map bill depth to the x-axis and map bill length to the y-axis. Represent each observation with a point and map species to the color of each point. Title the plot “Bill depth and length”
Start with the
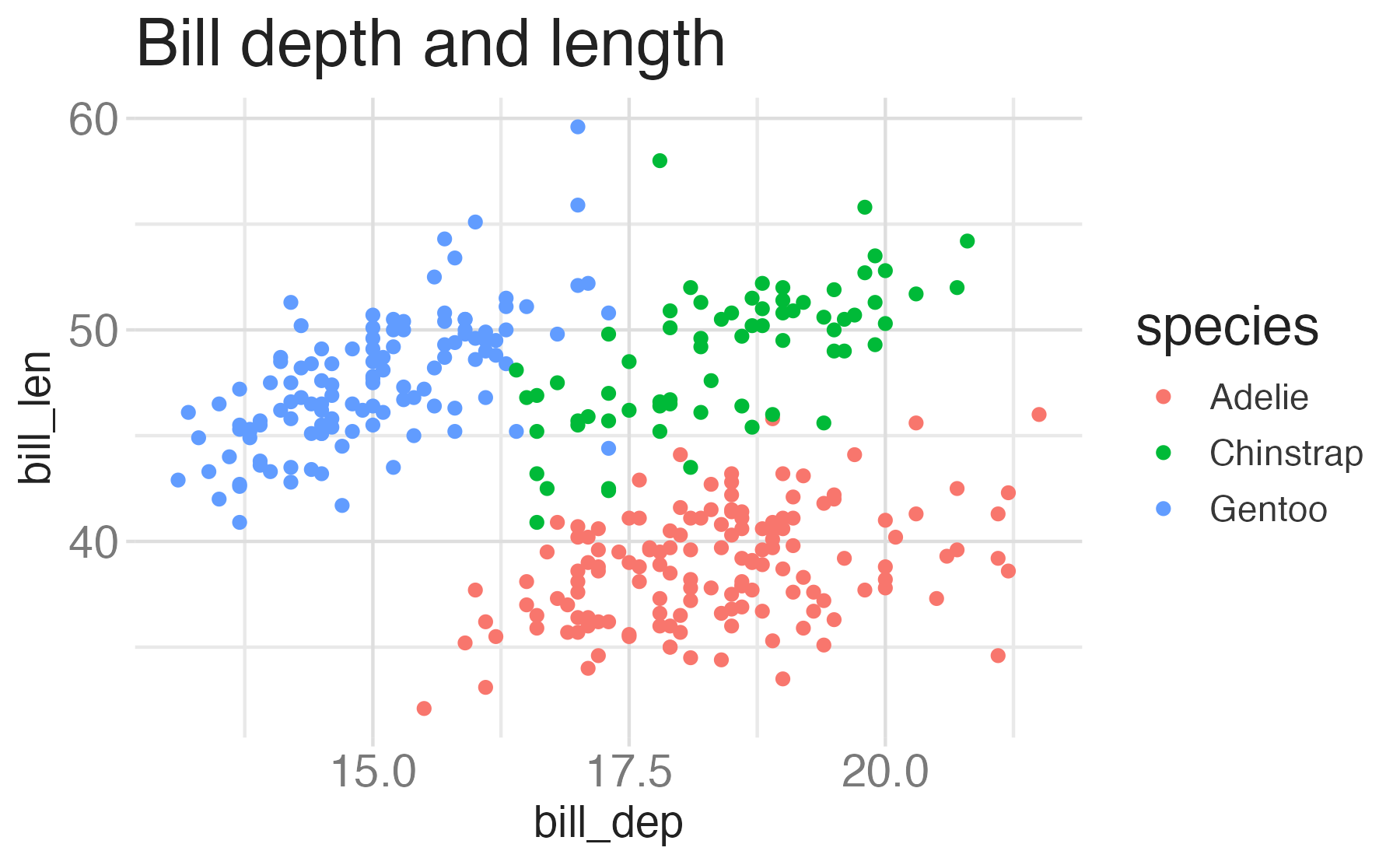
penguinsdata frame, map bill depth to the x-axis and map bill length to the y-axis. Represent each observation with a point and map species to the color of each point. Title the plot “Bill depth and length”, add the subtitle “Dimensions for Adelie, Chinstrap, and Gentoo Penguins”
Start with the
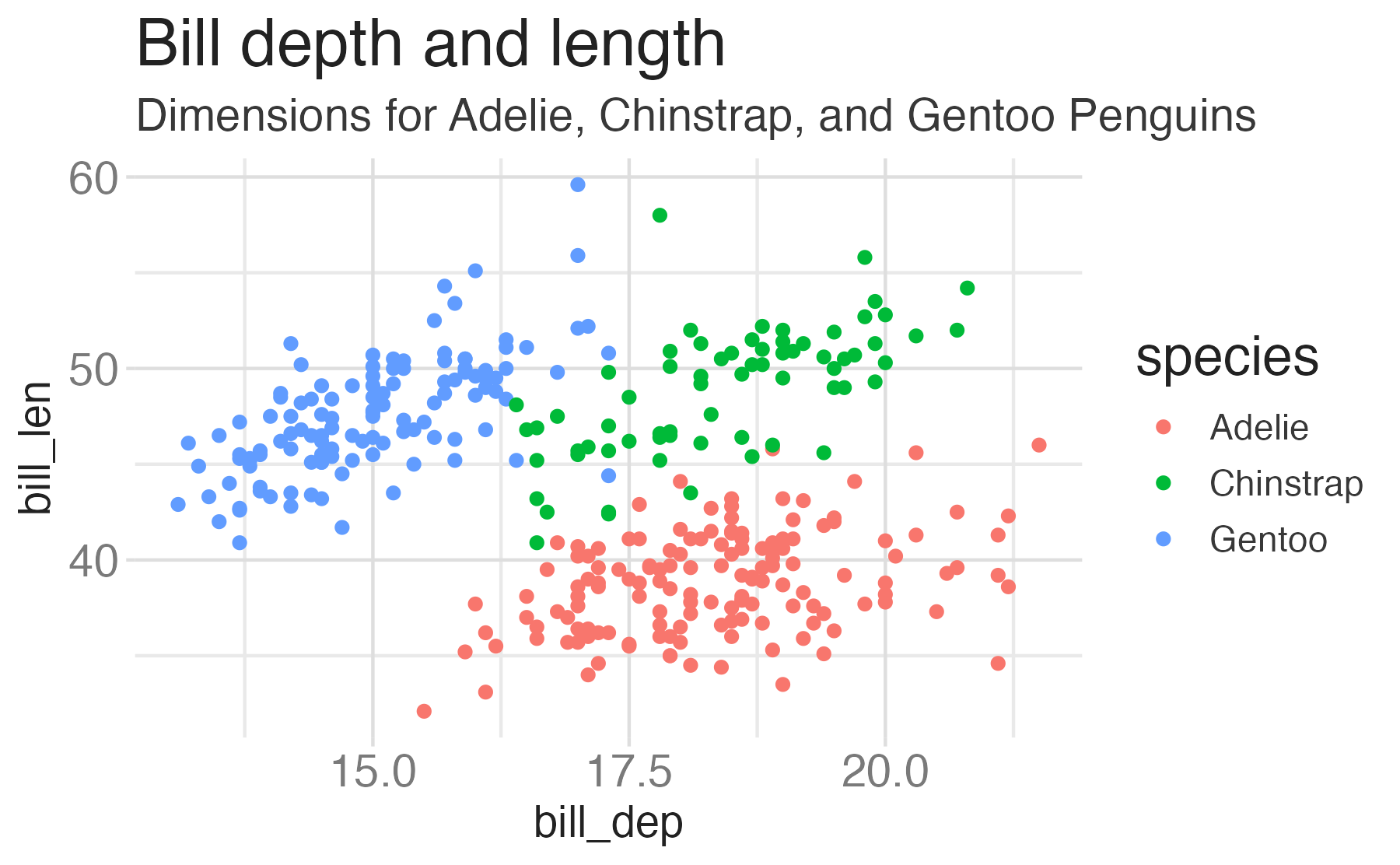
penguinsdata frame, map bill depth to the x-axis and map bill length to the y-axis. Represent each observation with a point and map species to the color of each point. Title the plot “Bill depth and length”, add the subtitle “Dimensions for Adelie, Chinstrap, and Gentoo Penguins”, label the x and y axes as “Bill depth (mm)” and “Bill length (mm)”, respectively
Start with the
penguinsdata frame, map bill depth to the x-axis and map bill length to the y-axis. Represent each observation with a point and map species to the color of each point. Title the plot “Bill depth and length”, add the subtitle “Dimensions for Adelie, Chinstrap, and Gentoo Penguins”, label the x and y axes as “Bill depth (mm)” and “Bill length (mm)”, respectively, label the legend “Species”
Start with the
penguinsdata frame, map bill depth to the x-axis and map bill length to the y-axis. Represent each observation with a point and map species to the color of each point. Title the plot “Bill depth and length”, add the subtitle “Dimensions for Adelie, Chinstrap, and Gentoo Penguins”, label the x and y axes as “Bill depth (mm)” and “Bill length (mm)”, respectively, label the legend “Species”, and add a caption for the data source.
ggplot(data = penguins,
mapping = aes(x = bill_dep,
y = bill_len,
color = species)) +
geom_point() +
labs(title = "Bill depth and length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Bill depth (mm)", y = "Bill length (mm)",
color = "Species",
caption = "Source: Palmer Station LTER / palmerpenguins package")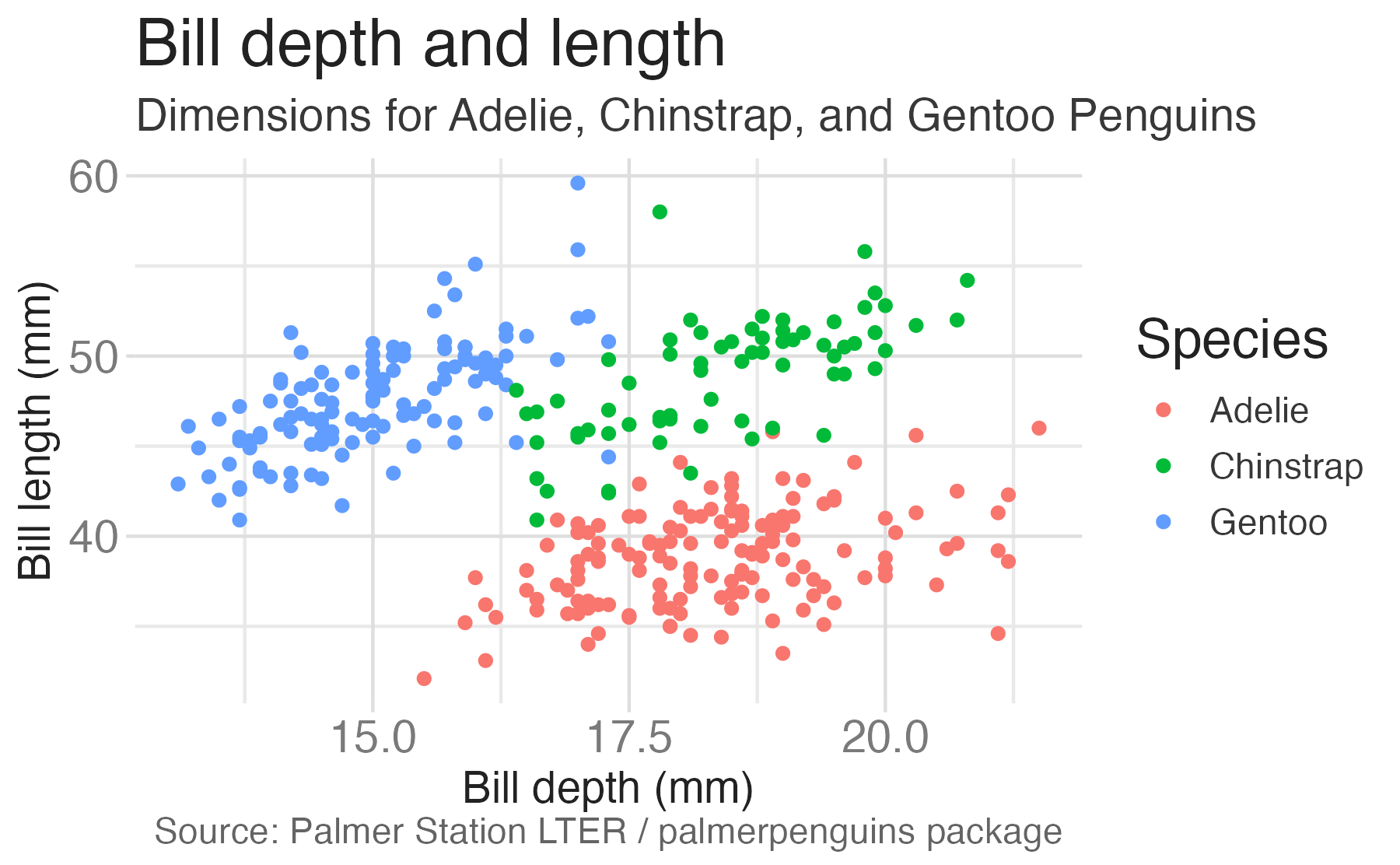
Start with the
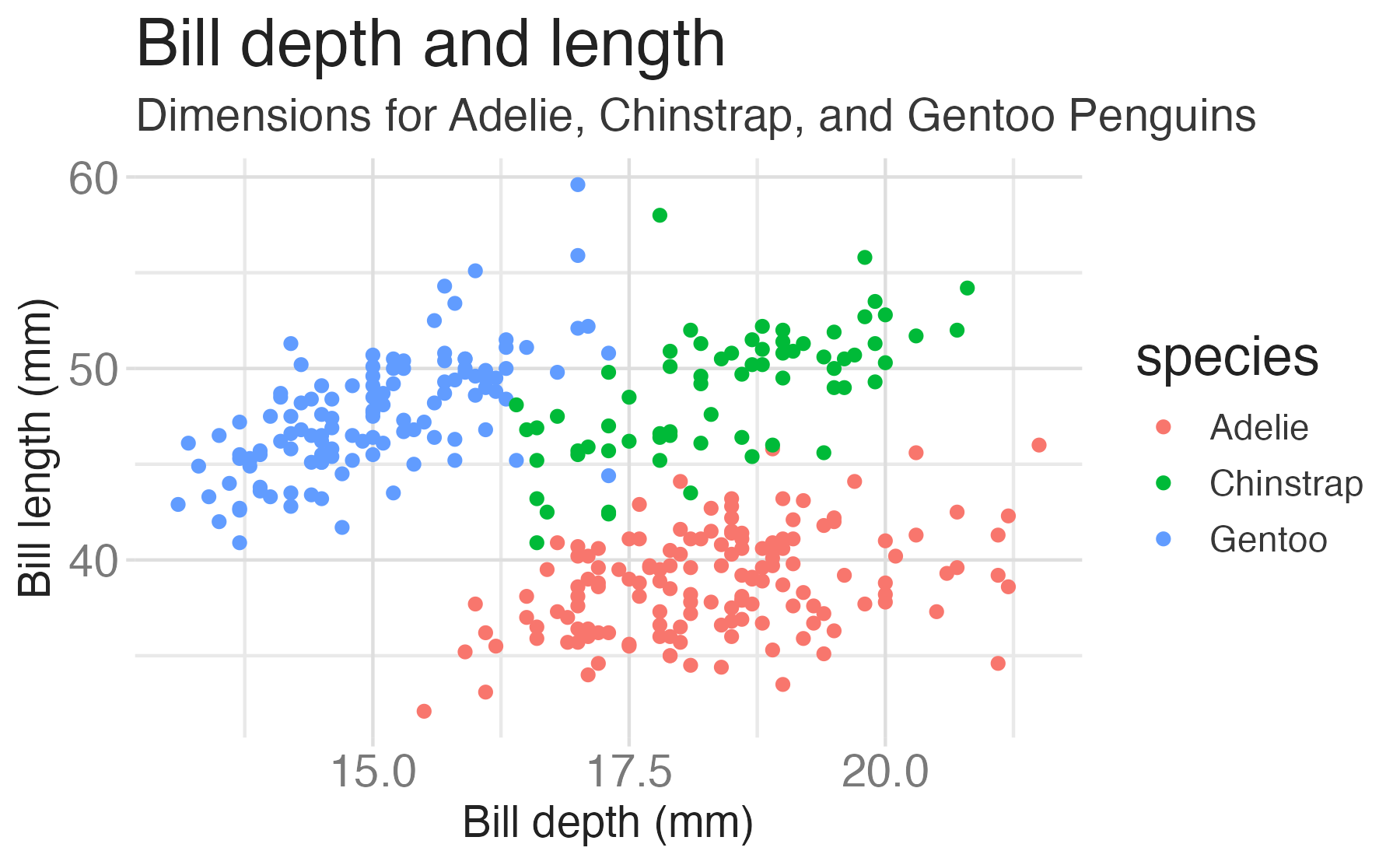
penguinsdata frame, map bill depth to the x-axis and map bill length to the y-axis. Represent each observation with a point and map species to the color of each point. Title the plot “Bill depth and length”, add the subtitle “Dimensions for Adelie, Chinstrap, and Gentoo Penguins”, label the x and y axes as “Bill depth (mm)” and “Bill length (mm)”, respectively, label the legend “Species”, and add a caption for the data source. Finally, use a discrete color scale that is designed to be perceived by viewers with common forms of color blindness.
ggplot(data = penguins,
mapping = aes(x = bill_dep,
y = bill_len,
color = species)) +
geom_point() +
labs(title = "Bill depth and length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Bill depth (mm)", y = "Bill length (mm)",
color = "Species",
caption = "Source: Palmer Station LTER / palmerpenguins package") +
scale_color_viridis_d()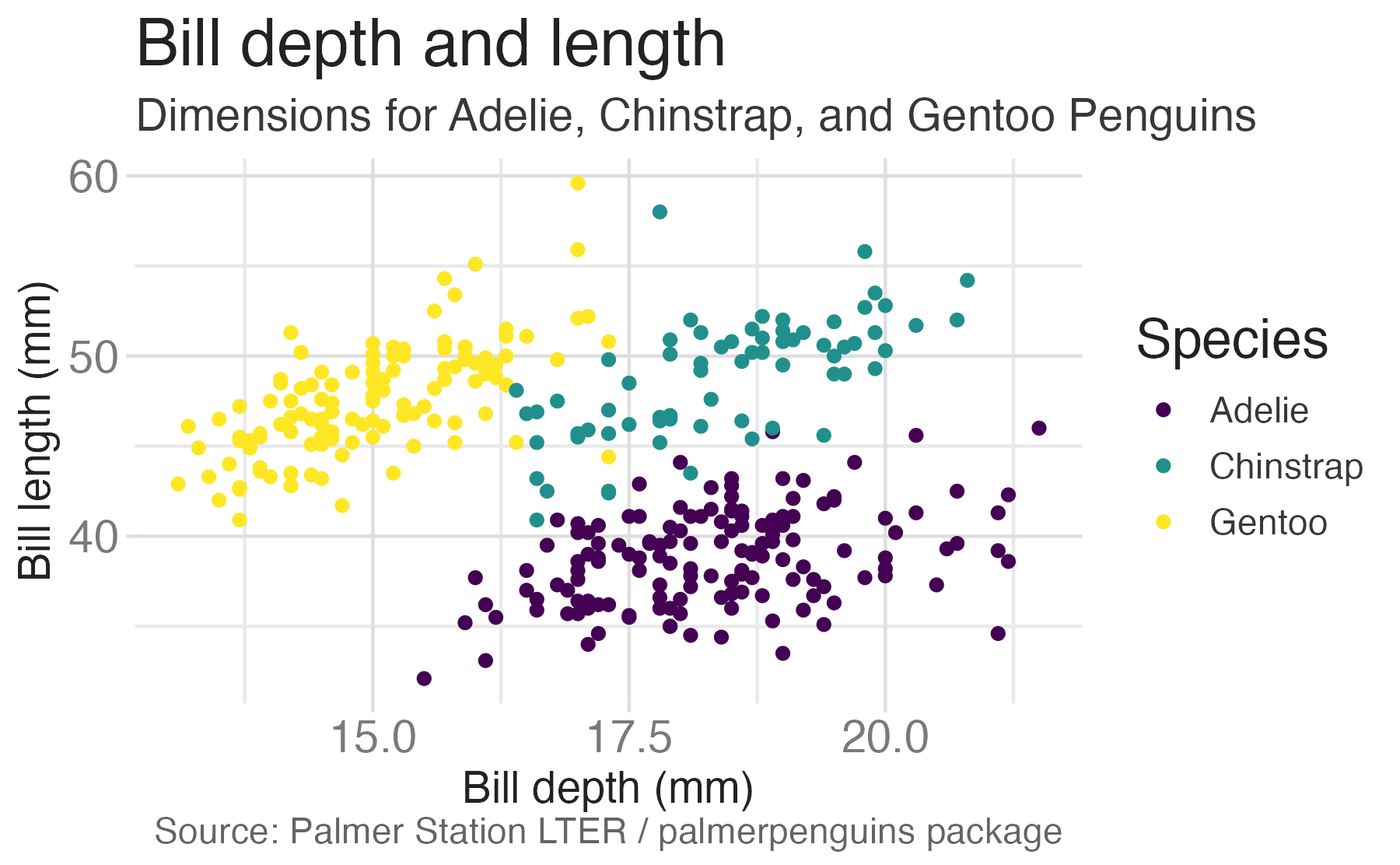
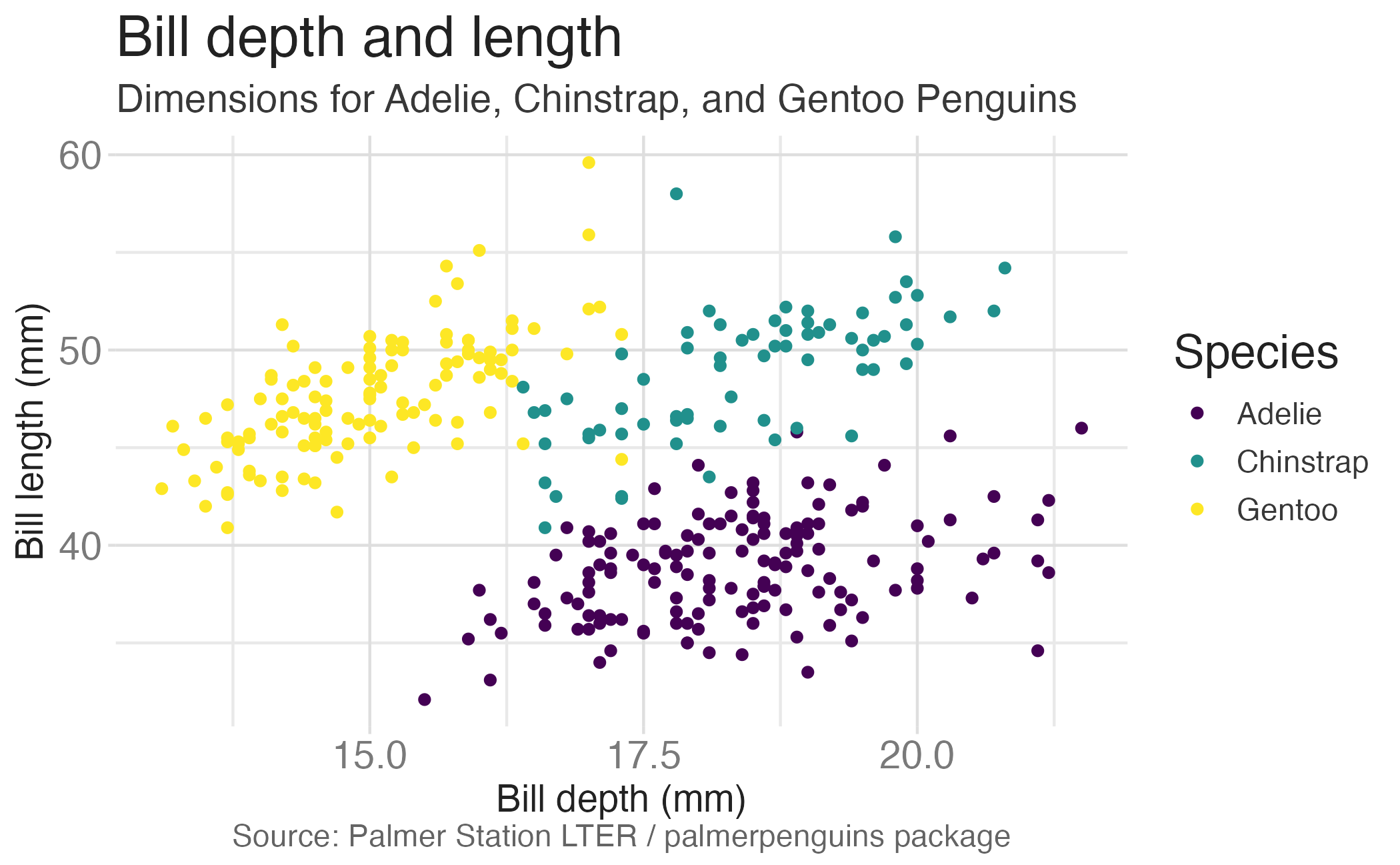
ggplot(data = penguins,
mapping = aes(x = bill_dep,
y = bill_len,
color = species)) +
geom_point() +
labs(title = "Bill depth and length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Bill depth (mm)", y = "Bill length (mm)",
color = "Species",
caption = "Source: Palmer Station LTER / palmerpenguins package") +
scale_color_viridis_d()Start with the penguins data frame, map bill depth to the x-axis and map bill length to the y-axis.
Represent each observation with a point and map species to the color of each point.
Title the plot “Bill depth and length”, add the subtitle “Dimensions for Adelie, Chinstrap, and Gentoo Penguins”, label the x and y axes as “Bill depth (mm)” and “Bill length (mm)”, respectively, label the legend “Species”, and add a caption for the data source.
Finally, use a discrete color scale that is designed to be perceived by viewers with common forms of color blindness.
Wrap up
Today’s tasks
- Log in to Cornell’s GitHub - you already have an account!
- Access Positron
- Complete the preparations for Thursday’s class